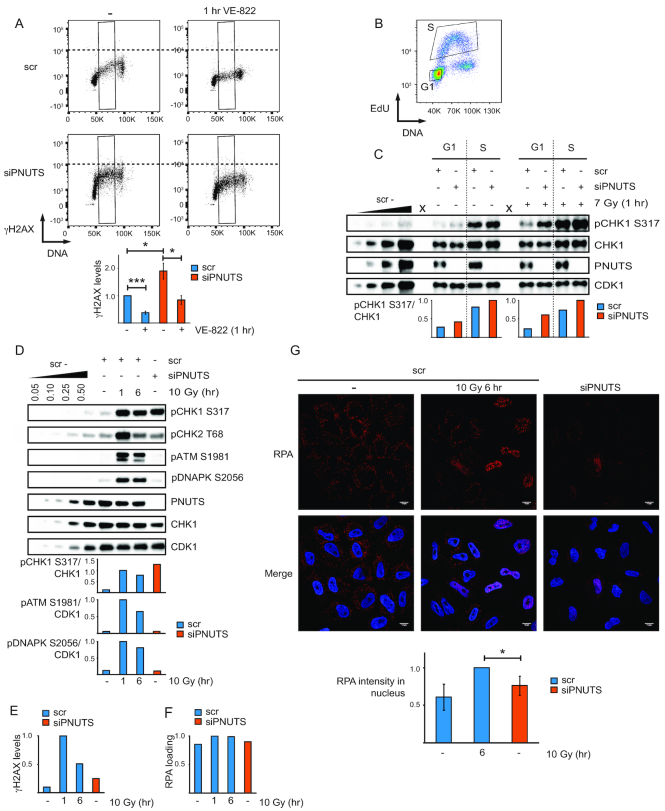
Figure 3.
High ATR signaling after PNUTS depletion is present in individual cells, does not correlate with DNA damage markers and can occur in G1-phase. (A) Flow cytometry charts showing γH2AX versus DNA staining of individual scr and siPNUTS transfected cells with and without VE-822 for 1 h. S-phase cells were gated based on DNA content as indicated (black boxes). Quantifications show average median γH2AX levels in S-phase (n = 3). *P < 0.05, ***P < 0.001 based on two-tailed two sample Student's t-test. (B) Cell sorting was performed by flow cytometry into G1- and S-phases based on EdU incorporation and DNA content as indicated. (C) Western blot analysis and quantifications of sorted (as in B) scr and siPNUTS transfected HeLa cells. Cells were harvested at 48 h after siRNA transfection, with and without IR (harvested at 1 h after 7 Gy). Irradiation was performed immediately prior to addition of EdU. One representative image is shown, with X indicating empty lanes. Quantifications were performed on images with different exposure times for the non-irradiated and irradiated samples (due to their different intensities), and normalized to the respective siPNUTS S-phase sample. The experiment was performed three times, two at 72 h and one at 48 h after siRNA transfection with similar results. (D) Western blot analysis of DNA damage markers for scr (without IR or 1 and 6 h after 10 Gy) and siPNUTS transfected cells 48 h after siRNA transfection. (E) Bar chart showing median levels of γH2AX from flow cytometry analysis from cells harvested in parallel with samples from the same experiment in D. The samples were barcoded with pacific blue and mixed prior to staining to minimise sample to sample variation. The experiment in (E) compared to (D) was performed two times with similar conditions and results. (F) Bar chart showing median levels of RPA loading from flow cytometry analysis of pre-extracted cells from the same experiment as in (D). Samples were barcoded as in (E). The experiment in (F) compared to (D) was performed three times with similar conditions and results. (G) Immunofluorescence analysis of pre-extracted cells treated as in (D), but harvested at 72 h after siRNA transfection. Bottom bar chart shows average intensity of nuclear RPA staining from three independent experiments. *P < 0.05 for using two-tailed one sample Student's t-test (to test if RPA values in siPNUTS sample was different than 1, which we had set scr 10 Gy 6 h sample to). >130 cells were scored per condition in total. Error bars represent SEM.