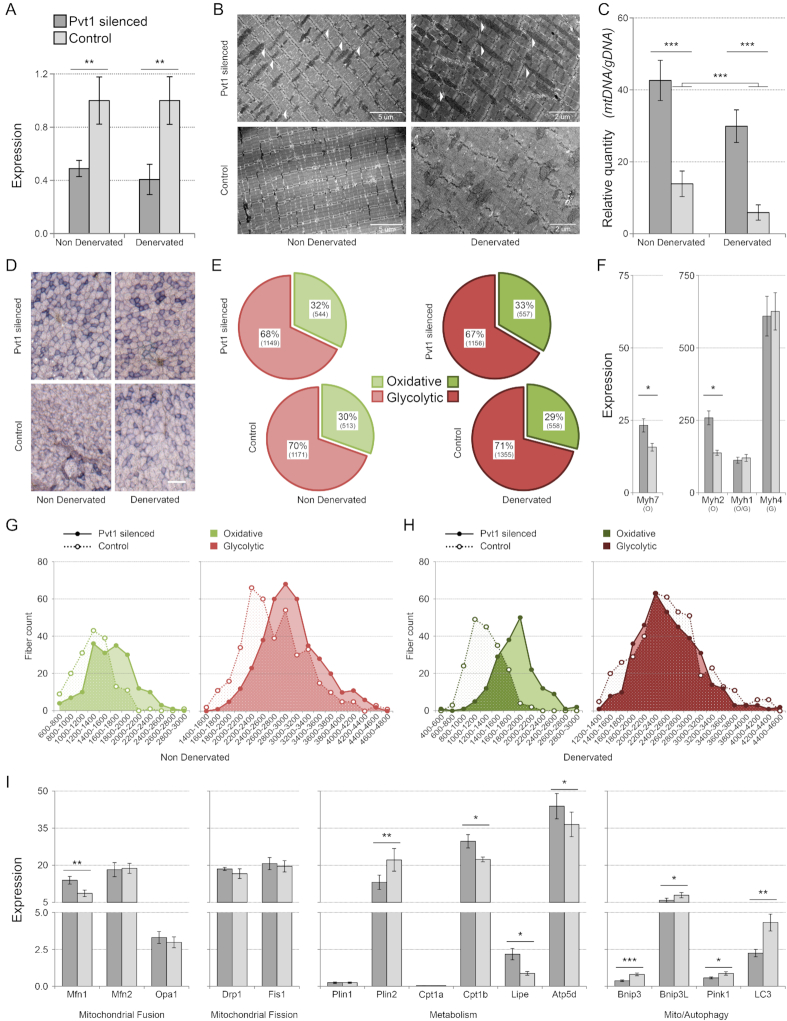
Figure 5.
In vivo analysis of Pvt1 function. (A) Relative expression of Pvt1 after silencing with GapmeRs. Standard deviation was calculated among three biological and two technical replicates per biological sample. (B) Electron microscopy of TA slices. White arrows indicate elongated mitochondria in samples where Pvt1 was low (in both normal and denervated muscles). When Pvt1 was down-regulated, the number of mitochondria increased. (C) Quantization of mitochondrial mass. Mitochondrial mass was expressed as relative quantity of mitochondrial DNA coding for Cox compared with nuclear DNA coding for Sdh. It increased when Pvt1 was down-regulated. The genomic region coding for TBP was used as reference. Four biological replicates were analyzed. Standard error is represented. (D) Representative images for Sdh staining in gastrocnemius muscles. Scale bar represents 100 μm. (E) Percentage of oxidative and glycolytic myofibers is represented. χ² tests reveal that the increase in oxidative fibers after Pvt1 silencing is not significant for non-denervated muscles (χ2 = 1.09, P-value = 0.296) while it is significant for denervated muscles (χ2 = 4.75, P-value = 0.029). About 3500 myofibers for each condition were counted. (F) Histograms represent expression values relative to control gene (Tbp), obtained by qPCR for transcripts for myosin heavy chain. After Pvt1 silencing, myosin heavy chain genes expressed in oxidative metabolic myofibers (Myh2 and Myh7) increased their expression. On the contrary, genes expressed in glycolytic myofibers (Myh1 expressed in oxidative/glycolytic and Myh4 in glycolytic) were not affected. Standard deviation was calculated from three biological and two technical replicates per biological sample. O = Oxidative; G = Glycolytic (G) Cross-section area (CSA) was measured for ∼1200 fibers. Values are expressed in μm2. Fibers were divided in groups having similar area and their frequency was plotted. In non-denervated muscles the CSA of oxidative and glycolytic myofibers slightly increased when Pvt1 was down-regulated. (H) In denervated muscles the CSA of oxidative myofibers does not decrease when Pvt1 was down-regulated while it was unaffected for glycolytic myofibers. (I) Histograms represent expression values relative to control gene (Tbp) and normalized against the average expression of the gene among samples. Among tested genes involved in the mitochondrial dynamics only Mfn1 was up-regulated after Pvt1 silencing. Metabolic related genes showed instead the up-regulation of Cpt1b, Lipe and Atp5d and the down-regulation of Plin2. All genes associated to mito/autophagy were down-regulated in muscle where Pvt1 was down-regulated. Standard deviation was calculated among three biological and two technical replicates Statistical significance was calculated according to Student’s t-test between the two groups considering a two-tailed distribution and samples having unequal variance. * P ≤ 5 × 10−2, ** P ≤ 1 × 10−2, *** P ≤ 1 × 10−3.