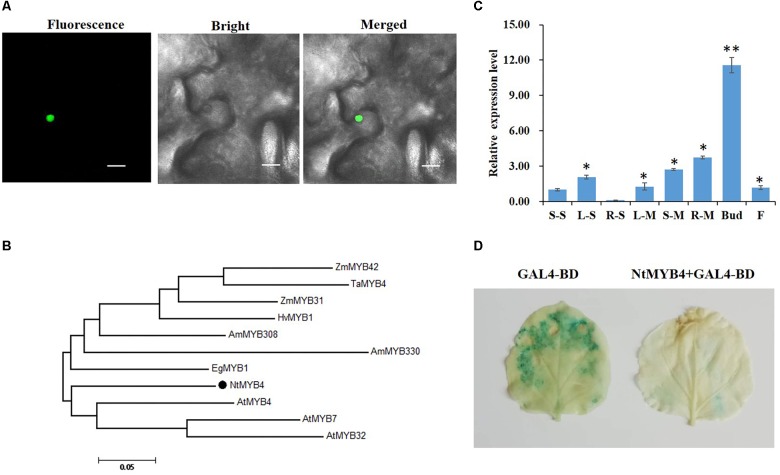
FIGURE 1.
NtMYB4 is a nuclear localized transcription repressor in tobacco. (A) Subcellular localization analysis of NtMYB4. An NtMYB4-GFP construct was transformed into leaves of N. benthamiana and examined by confocal laser scanning microscopy. A confocal micrograph is shown at the left (green fluorescent protein, GFP), the corresponding differential interference contrast (bright) image is in the middle, and the merged image is at the right. Bars, 20 μm. (B) Phylogenetic tree based on the amino acid sequences of NtMYB4 and 10 other R2R3 MYB subgroup 4 members. The NtMYB4 isolated in this study is highlighted by a black dot. Accession numbers for MYB subgroup 4 sequences are: AmMYB308 (JQ0960), AmMYB330 (P81395), AtMYB4 (AY519615), AtMYB7 (AEC06531), AtMYB32 (NP_195225), EgMYB1 (CAE09058), HvMYB1 (P20026), TaMYB4 (AAT37167), ZmMYB31 (CAJ42202), and ZmMYB42 (CAJ42204). (C) The expression pattern of NtMYB4 in tobacco root, stem, leaf flower and bud was detected by qRT-PCR. Expression data were detected in two representative growth stages of common tobacco, including the seedling (S) and mature (M) stages. Organs sampled included leaf (L), stem (S), root (R), flower (F). Asterisks in (C) indicate differences between tissues (∗P < 0.05, ∗∗P < 0.01). (D) UAS/GAL4-based transcriptional repression assay of NtMYB4 in N. benthamiana leaves. NtMYB4 repressed the 35S-driven GUS activity. Pairwise combinations of constructs as indicated were co-infiltrated into tobacco leaves and stained for GUS activity. The left leaf marked GAL4-BD is 35S-driven GAL4 DNA binding domain (BD) as control; the right leaf marked NtMYB4+GAL4-BD was 35S-driven full-length NtMYB4 fused to the Gal4-BD domain.