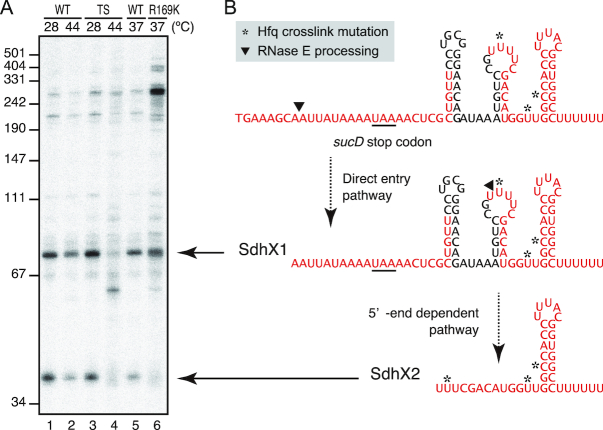
Figure 2.
SdhX is processed by RNase E. (A) Salmonella rne+ (WT: lanes 1–2) and rne3071 (TS: lanes 3–4) strains were grown to OD600 of 0.5 at 28°C, split into two flasks, and further incubated at either 28°C (lanes 1, 3) or 44°C (lanes 2, 4) for 30 min. Salmonella rne+ (WT: lane 5) and rneR169K (R169K: lane 6) strains were grown to OD600 of 0.5 at 37°C. The size is estimated by pUC19 MspI dsDNA fragments. (B) Predicted secondary structures of SdhX1 and SdhX2. Hfq-bound regions identified by CLIP-seq analysis (12) are indicated by red letters, and the mutations induced by crosslinking to Hfq are highlighted in yellow. See also Supplementary Figure S1.