Abstract
Twelve Ctenomyces (Arthrodermataceae, Onygenales) strains were obtained and identified during a survey of keratinophilic fungi in soils from China. We used molecular identification combined with morphological evidence to delimit species, circumscribing five species in the genus. Three new species are herein described: C.albussp. nov., C.obovatussp. nov. and C.peltricolorsp. nov. We also described, illustrated and compared the novel species with related species in the morphology.
Keywords: 3 new species, Filamentous fungi, Ctenomyces , Morphology, Multigene
Introduction
The genus Ctenomyces belongs in the family Arthrodermataceae in the order Onygenales (Wijayawardene et al. 2018) with C.serratus as the type species (Eidam 1880, Wijayawardene et al. 2017). Trichophytonlacticolor and Microsporummentagrophytes were transferred to Ctenomyces, being renamed C.lacticolor and C.mentagrophytes (Langeron and Milochevitch 1930). Subsequently, ten species (Trichophytondenticulatum, T.equinum, T.eriotrephon, T.farinulentum, T.felineum, T.griseum, T.persicolor, T.gypseumvar.radioplicatum, T.viannai and Epidermophytongypseum) were transferred to the genus (Nannizzi 1934). Thereafter, two new species, C.bossae and C.trichophyticus, were also described (Milochevitch 1935, Szathmáry 1960). Later studies showed that C.bossae was misnamed; C.trichophyticus was invalid; C.felineus was a synonym of C.serratus; C.persicolor was transferred to Nannizzia, and named as N.persicolor; C.mentagrophytes, C.equinus and C.eriotrephon were transferred to Trichophyton and named as T.mentagrophytes, T.equinum and T.eriotrephon, respectively; C.lacticolor and C.denticulatus were transferred to T.mentagrophytes; the remaining five species C.farinulentus, C.griseus, C.radioplicatus, C.viannai and C.gypseus were transferred back to Trichophyton and Epidermophyton, which were eventually regarded as invalid or unclear (Orr and Huehn 1963, de Hoog et al. 2000, de Hoog et al. 2017). Therefore, C.serratus was ultimately regarded as the only valid species within the genus (Orr and Huehn 1963).
The main diagnostic criteria of Ctenomyces (sensuOorschot 1980) are that conidia are verrucose, thick-walled, lightly pigmented, commonly with ampulliform swellings and mostly longer than 8 μm. Oorschot (1980) regarded Ctenomyces as a sexual morph of Myceliophthora. Furthermore, he transferred Chrysosporiumasperatum to Myceliophthora as M.vellerea in a taxonomic revision of Chrysosporium and allied genera. Myceliophthoravellerea was regarded as a synonym of the asexual morph of Ctenomycesserratus (Guarro et al. 1985, Chabasse 1988). Phylogenetic analyses, based on ITS rDNA, demonstrated Myceliophthoravellerea to be a synonym of Ctenomycesserratus (van den Brink et al. 2012). De Hoog et al. (2017) expanded the breadth and understanding of dermatophytes in Arthrodermatacea based on multi-locus molecular data and Ctenomycesserratus is used as the only previously validated species of the genus Ctenomyces.
Investigation of keratinophilic fungi has been given more attention in some countries (Anbu et al. 2004, Zarrin and Haghgoo 2011, Shadzi et al. 2002). Many researchers have shown that keratinophilic fungi distribution is closely related to human and animal activity (Sharma and Sharma 2010). Therefore, we conducted a survey of keratinophilic fungi in places with high human activity in Guizhou, Shanxi and Gansu provinces in China and isolated 12 strains. By combining the ITS sequence and a multi-gene phylogeny and the morphological characteristics, we identify and describe three new species and one new record of Ctenomyces from China.
Materials and methods
Isolates
Twelve Ctenomyces strains were obtained from soil samples collected in Guizhou, Shanxi and Gansu province of China using a baiting technique (Vanbreuseghem 1952). Sterile chicken feather and human hair were combined with the soil samples and the samples were placed in sterile Petri dishes, which were moistened with sterile distilled water. The baited soil sample Petri dishes were incubated at 25 °C for 1 month and remoistened as necessary. When fungal growth was observed, those feathers with fungal growth were mixed with 9 ml of sterile water in an Erlenmeyer flask and 1 ml of suspensions were evenly spread on plates containing Sabouraud’s dextrose agar (SDA) with chloramphenicol and cycloheximide medium. The plates were incubated at 25 °C. The pure culture were then transferred to potato dextrose agar (PDA) plates for purification, the isolates were inoculated to test-tube slants and stored at 4 °C.
All holotypes and isotypes were deposited in the Mycological Herbarium of the Institute of Microbiology, Chinese Academy of Sciences, Beijing, China (HMAS). Type strains and ex-type living cultures were deposited in the China General Microbiological Culture Collection Center (CGMCC) and the Institute of Fungus Resources, Guizhou University (GZUIFR). Taxonomic information of the new taxa was deposited in MycoBank (www.MycoBank.Org).
Morphology
Isolates were transferred to potato dextrose plates, incubated at 25 °C for 14 days and subjected to macroscopic examination. Fungal microscopic features were examined with a Nikon Ti-U microscope (Nikon, Japan) and photographed. Diagnostic features were then illustrated on the basis of these observations. Finally, the fungi were morphologically identified according to colony characteristics, conidiogenous structures and conidia (sensuOorschot 1980).
DNA extraction, PCR amplification, sequencing
Total genomic DNA was extracted from fresh sporulating cultures after 14 days at 25 °C using a Fungal DNA Mini Kit (Omega Biotech, Doraville, GA, USA) according to the manufacturer’s protocol and then stored at -20 °C. Three regions were amplified and sequenced, including the internal transcribed spacer (ITS) region using primers ITS1 and ITS4 (White et al. 1990); partial fragments of the RNA polymerase II largest subunit 2 (RPB2) gene region using primers 5F-Eur and 7cR-Eur (van den Brink et al. 2012); partial fragments of the translation elongation factor 1-alpha (EF1A) gene region using primers EF1-983F and EF1-2218R (van den Brink et al. 2012). The PCR mixture was prepared using a commercial kit (TSINGKE Biological Technology, Kunming, China) and contained 5 μl 10 × reaction buffer, 0.4 μl dNTPs (25μM), 0.2 μl T6 DNA polymerase (5 U/μl), 1 μl of each primer and 2 μl DNA template in a final volume of 25 μl. Reaction mixtures were pre-heated at 98 °C for 2 min and PCR was performed as follows: 30 cycles of 10 s at 98 °C, 10 s at 55 °C and 10 s 72 °C, with a final extension at 72 °C for 5 min and cooling at 4 °C. The PCR conditions were the same for all three markers. The resulting PCR products were sequenced by TSINGKE Biological Technology (Kunming, China) using the corresponding primers.
Phylogenetic analysis
Sequence data from the nine genera of Arthrodermataceae and Myceliophthoralutea sequences were used in the phylogenetic analysis. Details of newly generated and reference sequences retrieved from GenBank are listed in Table 1. Multiple sequence alignments for ITS, EF1A and RPB2 were achieved with MAFFT v.7.037b (Katoh and Standley 2013) and manually edited in the MEGA 6.06 (Tamura et al. 2013).
Table 1.
Strains included in the phylogenetic analysis.
| Taxa | Strain | GenBank accession | ||
|---|---|---|---|---|
| ITS | EF1A | RPB2 | ||
| Arthrodermauncinatum | IFO 31978 | JN134092 | KM678197 | |
| CBS 180.64 | MH858408 | KM678070 | ||
| Ctenomycesalbus | CGMCC 3.19232 T = GZUIFR-QL17.10 | MH793455 | MH801900 | MH801914 |
| CGMCC 3.18631 = GZUIFR-QL17.11 | MH793456 | MH801901 | MH801915 | |
| CGMCC 3.18632 = GZUIFR-QL17.12 | MH793457 | MH801902 | MH801916 | |
| C.obovatus | CGMCC 3.19225 T = GZUIFR-L15020 | MH793449 | MH801894 | MH801908 |
| CGMCC 3.19226 = GZUIFR-L15021 | MH793450 | MH801895 | MH801909 | |
| CGMCC 3.19227 = GZUIFR-L15022 | MH793451 | MH801896 | MH801910 | |
| C.peltricolor | CGMCC 3.19229 T = GZUIFR-C03010 | MH793458 | MH801903 | MH801917 |
| CGMCC 3.19230 = GZUIFR-C03011 | MH793459 | MH801904 | MH801918 | |
| CGMCC 3.19231 = GZUIFR-C03012 | MH793460 | MH801905 | MH801919 | |
| C.serratus | CBS 187.61 T | AJ877222 | ||
| CGMCC 3.18622 = ZUIFR-S37.1 | MH793452 | MH801897 | MH801911 | |
| CGMCC 3.18623 = GZUIFR-S37.2 | MH793453 | MH801898 | MH801912 | |
| CGMCC 3.18624 = GZUIFR-S37.3 | MH793454 | MH801899 | MH801913 | |
| C.vellereus | CBS 479.76 | HQ871797 | HQ871749 | HQ871840 |
| CBS 715.84 | HQ871795 | HQ871747 | HQ871841 | |
| Epidermophytonfloccosum | CBS 566.94 | MH862489 | ||
| CBS 457.65 | MH858667 | |||
| Guarromycesceretanicus | CBS 269.89 | MF926403 | ||
| Lophophytongallinae | CBS 244.66 | MH858789 | ||
| CBS 100083 | MF926355 | |||
| Microsporumaudouinii | CBS 545.93 T | KT155940 | ||
| CBS 404.61 | MF926387 | |||
| M.canis | CBS 496.86 T | KT155928 | ||
| CBS 101514 | KT155672 | |||
| M.ferrugineum | CBS 449.61 | KT155902 | ||
| CBS 373.71 | KT155886 | |||
| Nannizziafulva | CBS 168.64 | MH378229 | ||
| CBS 783.73 | MH378230 | |||
| N.persicolor | CBS 468.74 | AJ000615 | ||
| CBS 469.74 | AJ000614 | |||
| N.praecox | CBS 672.89 | MH378245 | ||
| CBS 673.89 | MH378246 | KM678113 | ||
| Paraphytoncookei | CBS 129.67 | MH858923 | KM678064 | |
| NBRC 7862 | JN134140 | KM678208 | ||
| P.cookiellum | CBS 101.83 T | KT155670 | ||
| CBS 102.83 | KT155674 | |||
| P.mirabile | CBS 129179 | MF926385 | ||
| CBS 124422 T | MF926384 | |||
| Trichophytonbenhamiae | CBS 808.72 | MH860614 | KM678050 | |
| CBS 807.72 | MH860613 | KM678118 | ||
| T.mentagrophytes | NBRC 5466 | JN134100 | KM678200 | |
| NBRC 5974 | JN134103 | KM678206 | ||
| T.rubrum | CBS 304.60 | AJ270807 | KM678081 | |
| CBS 734.88 | AJ270800 | KM678115 | ||
| T.simii | CBS 417.65 | MH858646 | KM678090 | |
| CBS 448.65 | MH858665 | KM678099 | ||
| Myceliophthoralutea | CBS 145.77 T | HQ871775 | HQ871722 | HQ871816 |
| MUCL 10070 | LK932701 | LK932710 | LK932724 | |
T= type strains, strain and sequences generated in this study are shown in bold.CBS: Westerdijk Fungal Biodiversity Institute, Utrecht, The Netherlands; CGMCC: China General Microbiological Culture Collection; NBRC: NITE Biological Resource Centre, Japan; IFO: Institute for Fermentation, Osaka, Yodogawa-ku, Osaka, Japan; GZUIFR: Guizhou University, Institute of Fungus Resources; MUCL: Belgian Co-ordinated Collections of Micro-organisms, Belgium.
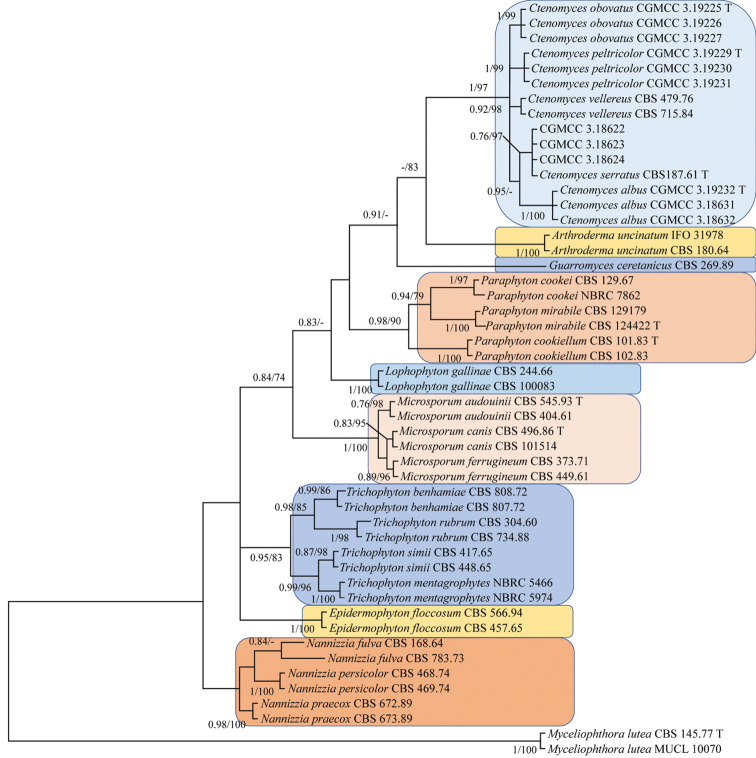
A total of 50 ITS sequences of 23 species and including Myceliophthoralutea (CBS 145.77 and MUCL 10070) as the outgroup taxon were used in the analysis. The data were analysed phylogenetically using Bayesian Markov chain Monte Carlo (MCMC) and maximum likelihood (ML). For the Bayesian analysis, two simultaneous Bayesian Inference (BI) Markov chain Monte Carlo runs were also executed for 10,000,000 generations, saving trees every 500 generations. Modeltest v3.7 suggested the GTR+I+G as the best-fit evolutionary model for dataset (Posada and Crandall 1988). After the BI analysis, each run was examined using the programme Tracer v1.5 (Drummond and Rambaut 2007) to determine whether the burn-in period was sufficient and to confirm that both runs had converged. ML analyses were performed using RAXML (Stamatakis and Alachiotis 2010) with the graphical user interface (GUI) (Silvestro and Michalak 2012) implementation and the GTRGAMMA model. The BI and ML analysis trees are available in TreeBASE (http://purl.org/phylo/treebase/phylows/study/TB2:S23736) and a consensus tree is presented in Figure 1.
Figure 1.
Phylogenetic tree of Arthrodermataceae based on the ITS dataset and Myceliophthoralutea (CBS 145.77 and MUCL 10070) as the outgroup taxon. Numbers at nodes are Bayesian posterior probabilities (left, BPP ≥0.75) and maximum likelihood bootstrap values (right, BS ≥70%).
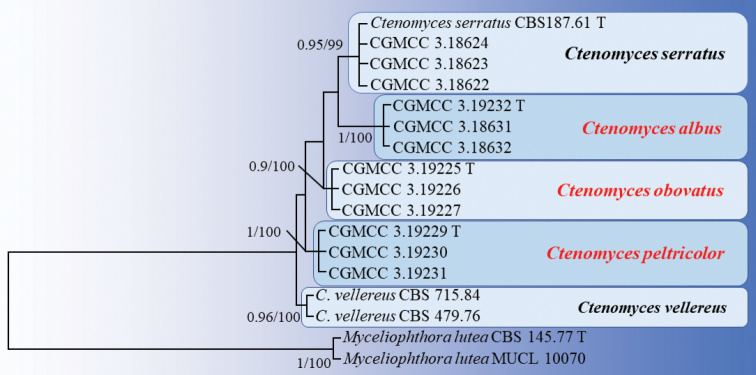
A concatenated dataset (ITS+EF1A+RPB2) of five Ctenomyces species and Myceliophthoralutea (CBS 145.77 and MUCL 10070) was assembled using SequenceMatrix v. 1.7.8 (Vaidya 2011). Concordance between genes was assessed with the ‘hompart’ command of PAUP4.0b10 (Swofford 2002). Maximum likelihood (ML) phylogenetic analyses of the datasets were performed using RAxML (Stamatakis and Alachiotis 2010) with the graphical user interface (GUI) (Silvestro and Michalak 2012) implementation and the General Time Reversible (GTR) model. Bootstrap analysis with 1,000 replicates was used to estimate nodal support. Two simultaneous Bayesian Inference Markov chain Monte Carlo runs were also executed for 10,000,000 generations, saving trees every 500 generations. Modeltest v3.7 suggested the GTR+I+G as the best-fit evolutionary model for the dataset (Posada and Crandall 1988). After the BI analyses, each run was examined using the programme Tracer v1.5 (Drummond and Rambaut 2007) to determine whether the burn-in period was sufficient and to confirm that both runs had converged. The BI and ML analyses trees are available in TreeBASE (http://purl.org/phylo/treebase/phylows/study/TB2:S23736) and a consensus tree is presented in Figure 2.
Figure 2.
Phylogenetic tree of Ctenomyces based on the ITS+EF1A+RPB2 dataset and Myceliophthoralutea (CBS 145.77 and MUCL 10070) as the outgroup taxon. Numbers at nodes are Bayesian posterior probabilities (left, BPP ≥0.9) and maximum likelihood bootstrap values (right, ≥95%).
Results
Phylogeny analysis
The ITS sequence alignment comprised 50 strains of 23 species (Table 1). The final dataset comprised 609 characters after alignment, which included nine genera of Arthrodermataceeae: Arthroderma, Ctenomyces, Epidermophyton, Guarromyces, Lophophyton, Microsporum, Nannizzia, Paraphyton and Trichophyton and Myceliophthoralutea (CBS 145.77 and MUCL 10070). No significant differences in topology were observed between the BI and ML phylogenies. The phylogenies show that each genus to cluster into the expected subclades (Figure 1). The Ctenomyces strains all cluster in a single clade with good nodal support (BPP 1, BS 97%) and were divided into four subclades, including the C.vellereus and the type species, C.serratus. All species of Ctenomyces clustered in separate well-supported subclades comprising C.serratus (BPP 0.76, BS 97%), C.vellereus (BPP 0.92, BS 98%), C.albus (BPP 1, BS 100%), C.obovatus (BPP 1, BS 99%) and C.peltricolor (BPP 1, BS 99%).
The combined ITS+EF1A+RPB2 sequence alignment comprised 17 taxa of six species within Ctenomyces and Myceliophthora (Table 1). The total length of sequences were 1,719 (ITS: 453 bp, EF1A: 728 bp, RPB2: 538 bp) characters and M.lutea (CBS 145.77 and MUCL 10070) was the designated outgroup taxon. The BI and ML analysis yielded congruent tree topology (Figure 2). The phylogenies show that each genus was sorted into expected subclades. Particularly, C.vellereusCBS 479.76 and CBS 715.84 strains cluster together with strong support values (BPP 0.96, BS 100%) and were separate from C.serratus (CBS 187.71, CGMCC 3.18622, CGMCC 3.18623 and CGMCC 3.18624) clustering together with strong support (BPP 0.95, BS 99%). In addition, our proposed three new species: C.albus, C.obovatus and C.peltricolor, had high support as a single subclade 1/100%, 0.9/100% and 1/100%, respectively.
Taxonomy
Ctenomyces albus
Y.F. Han, Z.Q. Liang & Z.Y. Zhang sp. nov.
827872
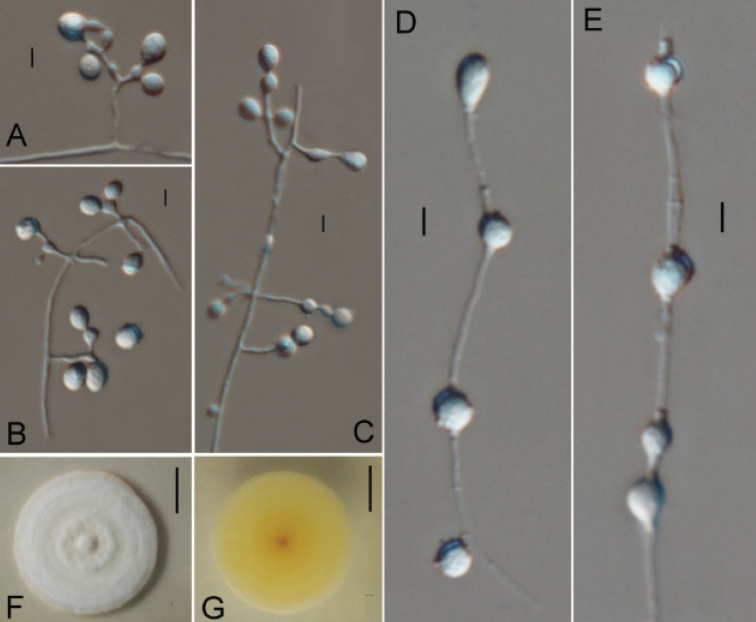
Figure 3.
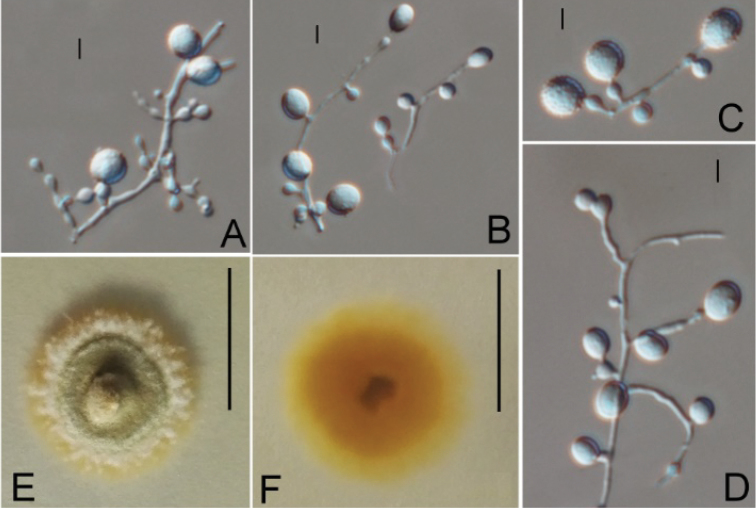
Ctenomycesalbus (from ex-holotype strain CGMCC 3.19232). A–C Conidiogenous structures and conidia D, E Intercalary conidia F, G Colony on PDA at day 14. Scale bars: 10 µm (A–E); 10 mm (F, G).
Holotype.
CHINA, Guizhou Province, on soil, Sept. 2016, Z.Y. Zhang (HMAS 255389, holotype, ex-type culture CGMCC 3.19232).
Paratypes.
CHINA, Guizhou Province, on soil, Sept. 2016, Z.Y. Zhang, dried cultures HMAS 255442 and HMAS 255443, isolates CGMCC 3.18631 (GZUIFR-QL17.11) and CGMCC 3.18632 (GZUIFR-QL17.12).
Etymology.
Referring to the white colony.
Description.
Aerial hyphae hyaline, smooth, septate, branched, 1.1–2.4 μm wide; racquet hyphae absent. Terminal and lateral conidia borne on hyphae, short protrusions, side branches or an ampulliform swelling. Conidia solitary or in series of up to 2–3 conidia connected by short and slim hypha, ellipsoid, smooth- or rough-walled, verrucose, 12.8–18.6 × 10.8–14.7 μm (x‒ = 15.4 × 12.5 μm, n=15). Intercalary conidia present, subglobose or ellipsoidal, smooth- or rough-walled, 13.1–16.9 × 11.2–14.4 μm (x‒ = 14.5 × 12.6 μm, n=15).
Culture characteristics.
Colonies on PDA growing in the dark reaching 32 mm diam. in 14 d at 25 °C, white, short fluffy to powdery, appearing some annulations, rounded, margin regular and defined. Reverse yellowish.
Notes.
Ctenomycesalbus is distinct from other species as it is the only species with intercalary conidia in the genus. In addition, our ITS and polygenic phylogeny showed that three isolates of C.albus were in a clade sister to C.serratus (Figures 1, 2) and clearly separate from other species. Following Jeewon and Hyde’s (2016) guidelines on new species delimitation, there were 37 bp (base pair) differences amongst 508 nucleotides ITS sequences between the isolate CGMCC 3.19232 and C.serratusCBS 187.61 (only ITS sequence data are available, EF1A and RPB2 are lacking), which also supports them as distinct different species. Therefore, we introduce C.albus sp. nov. in this study.
Ctenomyces obovatus
Y.F. Han, Z.Q. Liang & Z.Y. Zhang sp. nov.
827869
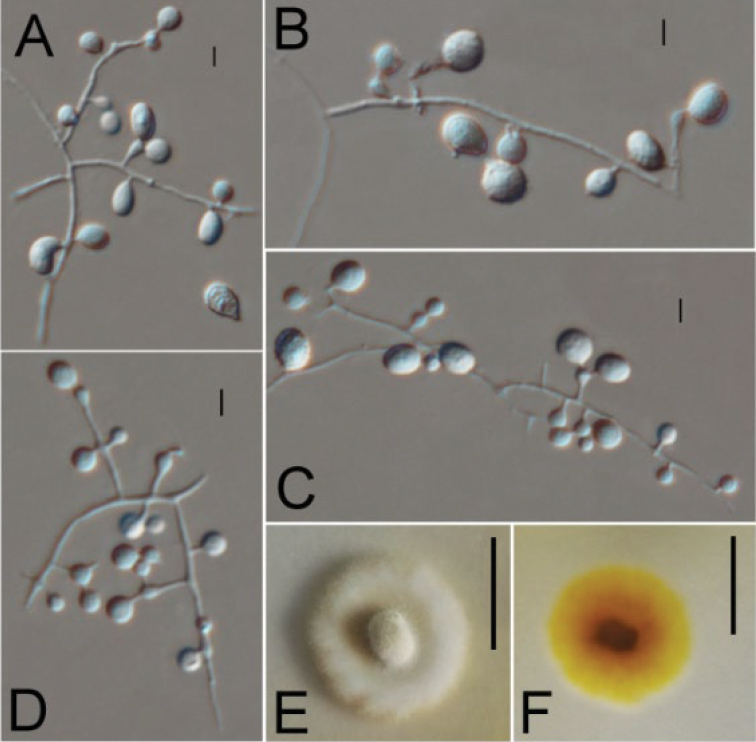
Figure 4.
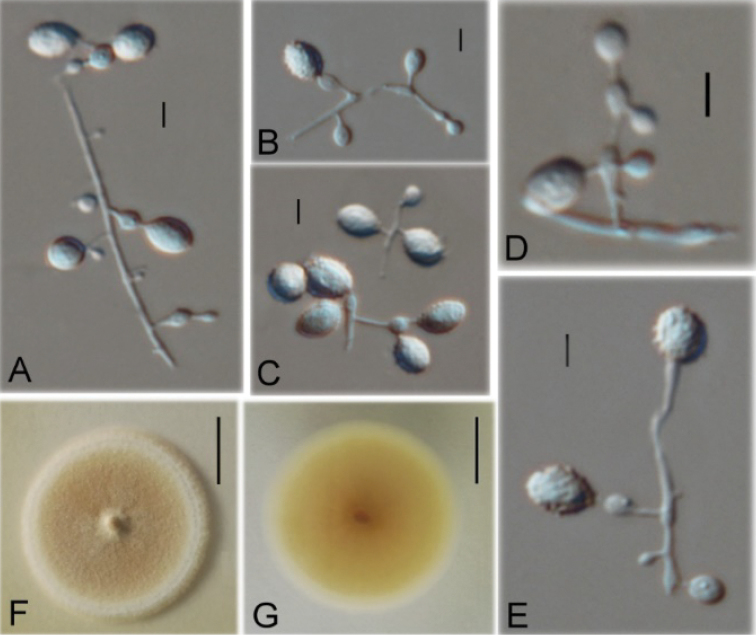
Ctenomycesobovatus (from ex-holotype strain CGMCC 3.19225). A–D Conidiogenous structures and conidia E, F Colony on PDA at day 14. Scale bars: 10 µm (A–D); 10 mm (E, F).
Holotype.
CHINA, Shanxi Province, on soil, Nov. 2017, Z.Y. Zhang (HMAS 255446, holotype, ex-type culture CGMCC 3.19225).
Paratypes.
CHINA, Shanxi Province, on soil, Nov. 2017, Z.Y. Zhang, dried cultures HMAS 255447 and HMAS 255448, isolates CGMCC 3.19226 (GZUIFR-L15021) and CGMCC 3.19227 (GZUIFR-L15023).
Etymology.
Referring to the obovoid conidia.
Description.
Aerial hyphae hyaline, smooth, septate, abundant branched, 1.2–2.4 μm wide; racquet hyphae absent. Terminal and lateral conidia borne on hyphae, short protrusions, side branches or an ampulliform swelling. Conidia solitary or in series of up to 2 conidia, ellipsoidal, obovoid, smooth- or rough-walled, verrucose, spinate, 10.3–17.3 × 9.7–10.5 μm (x‒ = 14.5 × 10 μm, n=15). Intercalary conidia absent.
Culture characteristics.
Colonies on PDA growing in the dark reaching 14–15 mm diam. in 14 d at 25 °C, yellowish, white in the margin; fluffy; rounded, margin regular. Reverse brown.
Notes.
Ctenomycesobovatus resembles C.vellereus in conidia size and conidiogenous cells. However, C.obovatus is the only species that produces obovoid conidia in this genus. Furthermore, our ITS and multigene phylogeny shows that three isolates of C.obovatus formed a single clade separate from other species (Figure 1), which indicates that C.obovatus is a new species.
Ctenomyces peltricolor
Y.F. Han, Z.Q. Liang & Z.Y. Zhang sp. nov.
827873
Figure 5.
Ctenomycespeltricolor (from ex-holotype strain CGMCC 3.19229) A–D Conidiogenous structures and conidia E, F Colony on PDA at day 14. Scale bars: 10 µm (A–D); 10 mm (E, F).
Holotype.
CHINA, Gansu Province, on soil, Nov. 2017, Z.Y. Zhang (HMAS 255387, holotype, ex-type culture CGMCC 3.19229).
Paratypes.
CHINA, Gansu Province, on soil, Nov. 2017, Z.Y. Zhang, dried cultures HMAS 255439 and HMAS 255440, isolates CGMCC 3.19230 (GZUIFR-C03011) and CGMCC 3.19231 (GZUIFR-C03012).
Etymology.
Referring to the pewter colony.
Description.
Aerial hyphae hyaline, smooth, septate, branched, 1.2–3.3 μm wide; racquet hyphae absent. Terminal and lateral conidia borne on hyphae, short protrusions, side branches or an ampulliform swelling. Conidia solitary, usually only 1 borne on ampulliform swellings; subglobose or globose; smooth- or rough-walled, verrucose, spinate, 8.3–20.2 μm (x‒ = 15.5 μm, n=15). Intercalary conidia absent.
Culture characteristics.
Colonies on PDA growing in the dark reaching 12 mm diam. in 14 d at 25 °C, pewter at the centre, white in the margin; powdery to floccose at the centre, fluffy in the margin; appearing a circle of annulation; rounded, margin regular. Reverse brown at the centre, yellowish in the margin.
Notes.
Ctenomycespeltricolor is distinct from other species in its single conidia borne on ampulliform swellings and colony colour. Phylogenetically, the ITS-based phylogenetic analysis (Figure 1) showed that three isolates of C.peltricolor cluster in a single clade (BPP 1, BS 100%), consistent with the results of multigene phylogenetic analysis (BPP 1, BS 100%) (Figure 2). Therefore, based on both morphological and phylogenetic evidence, C.peltricolor was identified as a new species of Ctenomyces.
Ctenomyces serratus
Eidam, in Beitrag zur Kenntnis der Gymnoasceen, Beiträge zur Biologie der Pflanzen 3: 267–305 (1880)
827871
Figure 6.
Ctenomycesserratus (from strain CGMCC 3.18622) A–E Conidiogenous structures and conidia F, G Colony on PDA at day 14. Scale bars: 10 µm (A–E); 10 mm (F, G).
Description.
Aerial hyphae hyaline, smooth, septate, branched, 0.9–3.3 μm wide; racquet hyphae absent. Terminal and lateral conidia borne on hyphae, short protrusions, side branches or an ampulliform swelling; ampulliform swelling solitary or 2 in series. Conidia solitary or in series of up to 2–3 conidia connected by short and slim hypha, mostly ellipsoidal, sometimes subglobose; smooth- or rough-walled, verrucose, spinate, 11.5–21.9 × 8–15.2 μm (x‒ = 18.5 × 13.2 μm, n=15). Intercalary conidia absent.
Culture characteristics.
Colonies on PDA growing in the dark reaching 30 mm diam. in 14 d at 25 °C, brown, white in the margin, floccose at the centre, short fluffy in other part, appearing obvious annulation; rounded, margin regular and defined. Reverse yellowish.
Specimens examined.
CHINA, Guizhou Province, on soil, Sept. 2016, Z.Y. Zhang, dried cultures HMAS 255390, HMAS 255444 and HMAS 255445, isolates CGMCC 3.18622 (GZUIFR-S37.1), CGMCC 3.18623 (GZUIFR-S37.2) and CGMCC 3.18624 (GZUIFR-S37.3).
Known distribution.
Currently known from Australia, England, India, Argentina, Germany (sensuOorschot 1980) and China (this study).
Notes.
The Australian collection of C.serratus (CBS 187.61) lacks racquet hyphae, conidia occur occasionally on short protrusions and usually 1–2 are borne on ampulliform swellings, solitary or in series of up to 3 conidia separated by short, narrow hyphal segments, initially subhyaline, thin- and smooth-walled, soon becoming reddish-brown, verrucose and thick-walled, ellipsoid, 5–23 × 3.5–12 µm, mature conidia usually 12–23 × 10.5–12 µm, with narrow basal scars (approx. 1 µm) (sensuOorschot 1980). The characteristic features data from the China collections matched rather well with the original description of C.serratus reported from Australia. Phylogenetically, our isolates CGMCC 3.18622, CGMCC 3.18623 and CGMCC 3.18624 shared a close relationship with C.serratus (Figures 1, 2). Therefore, the isolates CGMCC 3.18622, CGMCC 3.18623 and CGMCC 3.18624 were identified as C.serratus which was new to China.
Discussion
Members of the family Arthrodermataceae (Onygenales) were common in nature, mostly found as saprobes in soil on keratin-rich substrates or associated with vertebrate causing dermatophytosis and other infections. The widely accepted morphology-based taxonomy of dermatophytes in the genera Trichophyton, Microsporum and Epidermophyton was established by Emmons (Emmons 1934). There are three common ecological groups, anthropophilic, zoophilic or geophilic (de Hoog et al. 2017). Trichophyton and Epidermophyton usually belonged to anthropophiles or zoophilic taxa, Microsporum were considered to zoophilic, Arthroderma was geophilic. Some species cannot be clearly attributed to one of these groups due to insufficient data. Geophilic dermatophytes have their reservoir in the soil around burrows of specific terrestrial mammals, feeding on keratinous debris. Hence, the difference between geophilic and zoophilic dermatophytes is not always well-resolved. The genus of Ctenomyces is usually saprobic or closely related to keratin-rich substrates (Wijayawardene et al. 2017, Deshmukh et al. 2018). In addition, these strains of our study were isolated by using hair and feathers as baiting material. Therefore, we infer the genus Ctenomyces to be geophilic and/or zoophilic.
Phylogenetic studies based on the ITS (Graser et al. 2008), partial LSU, the ribosomal 60S protein, partial β-tubulin and translation elongation factor 3 for Arthrodermataceae have shown that the genus Trichophyton was polyphyletic and resulted in establishing nine genera, i.e. Arthroderma, Ctenomyces, Epidermophyton, Guarromyces, Lophophyton, Microsporum, Nannizzia, Paraphyton and Trichophyton and it suggested that ITS was the optimal barcoding marker (de Hoog et al. 2017). Therefore, we selected the ITS sequences for phylogenetic analysis of Arthrodermataceae in this study and our results are consistent with the previous studies (de Hoog et al. 2017). Ctenomycesvellereus (strains CBS 479.76 and CBS 715.84) had ITS, EF1A and RPB2 sequence data. Hence, we selected the ITS, EF1A and RPB2 genes for phylogenetic analysis of Ctenomyces. The results show that the ITS-based and multigene-based phylogenetic analyses have similar results. Although our study revealed that Ctenomyces was closely related to Arthroderma in Arthrodermataceae, Onygenales, the species of Ctenomyces nearly all produce ampulliform swellings, a feature absent in Arthroderma.
Guarro et al. (1985), Chabasse (1988) and van den Brink et al. (2012) proposed that C.vellereus was a synonym of C.serratus, however, these two species have several different characters, the conidia of C.serratus are ellipsoid, 12–23 × 10.5–12 µm, while those of C.vellereus are subglobose, pyriform or ellipsoid, 4–13 × 3–9 μm (sensuOorschot 1980). Van den Brink et al. (2012) conducted phylogenetic analysis of ITS sequences including only one sequence of C.serratus and three sequences of C.vellereus. This study used more ITS sequences in Ctenomyces for phylogenetic analysis and multigenic phylogeny analysis showed that they were two different species. In our phylogenetic tree, C.albus, C.obovatus, C.peltricolor, C.serratus and C.vellereus were grouped in five clear clades with good supported value and they are distinct from each other. Thus, based on the present molecular phylogeny, derived from nuclear and ribosomal DNA sequence data, together with morphological evidence, three distinct new Ctenomyces species, C.albus, C.obovatus, C.peltricolor and one new record, C.serratus, were proposed.
Key to the species of the genus Ctenomyces
| 1 | Intercalary conidia absen | 2 |
| – | Intercalary conidia present, subglobose or ellipsoidal | C.albus |
| 2 | Mostly 1–2 conidia borne on ampulliform swellings | 3 |
| – | Usually only 1 conidia borne on ampulliform swellings | C.peltricolor |
| 3 | Conidia less than 20 μm long | 4 |
| – | Conidia more than 20 μm long | C.serratus |
| 4 | Conidia obovoid or ellipsoidal | C.obovatus |
| – | Conidia subglobose, pyriform or ellipsoid | C.vellereus |
Supplementary Material
Acknowledgements
This work was financially supported by Ministry of Science and Technology of China (2013FY110400), the National Natural Science Foundation of China (31460010, 31860002), the Major Project of Guizhou Province, China (Qian Ke He Major Project [2016] 3022-07) and Construction Program of Biology First-class Discipline in Guizhou (GNYL [2017] 009). We thank Steven M. Thompson, from Liwen Bianji, Edanz Editing China (www.liwenbianji.cn/ac), for editing the English text of a draft of this manuscript.
Citation
Zhang Z-Y, Han Y-F, Chen W-H, Liang Z-Q (2019) Phylogeny and taxonomy of three new Ctenomyces (Arthrodermataceae, Onygenales) species from China. MycoKeys 47: 1–16. https://doi.org/10.3897/mycokeys.47.30740
References
- Anbu P, Hilda A, Gopinath SCB. (2004) Keratinophilic fungi of poultry farm and feather dumping soil in Tamil Nadu, India. Mycopathologia 158: 303–309. 10.1007/s11046-004-3465-1 [DOI] [PubMed] [Google Scholar]
- Chabasse D. (1988) Taxonomic study of keratinophilic fungi isolated from soil and some mammals in France. Mycopathologia 101: 133–140. 10.1007/BF00437030 [DOI] [PubMed] [Google Scholar]
- de Hoog GS, Dukik K, Monod M, Packeu A, Stubbe D, Hendrickx M, Kupsch C, Stielow JB, Freeke J, Göker M, Rezaei-Matehkolaei A, Mirhendi H, Gräser Y. (2017) Toward a novel multilocus phylogenetic taxonomy for the dermatophytes. Mycopathologia 182: 5–31. 10.1007/s11046-016-0073-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Hoog GS, Guarro J, Gené J, Figueras MJ. (2000) Atlas of Clinical Fungi (2nd edn). American Society for Microbiology Press, Washington, DC.
- Deshmukh SK, Verekar SA, Chavan YG. (2018) Keratinophilic fungi from the vicinity of salt pan soils of Sambhar lake Rajasthan (India). Journal de Mycologie Médicale 28: 457–461. 10.1016/j.mycmed.2018.06.002 [DOI] [PubMed] [Google Scholar]
- Drummond A, Rambaut A. (2007) BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evolutionary Biology 7: 214–221. 10.1186/1471-2148-7-214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eidam E. (1880) Beitrag zur Kenntnis der Gymnoasceen. Beiträge zur Biologie der Pflanzen 3: 267–305. [Google Scholar]
- Emmons CW. (1934) Dermatophytes: natural groupings based on the form of the spores and accessory organs. Archives of Dermatology and Syphilology 30: 337–362. 10.1001/archderm.1934.01460150003001 [DOI] [Google Scholar]
- Guarro J, Punsola L, Cano J. (1985) Myceliophthoravellerea (Chrysosporiumasperatum) anamorph of Ctenomycesserratus. Mycotaxon 23: 419–427. [Google Scholar]
- Jeewon R, Hyde KD. (2016) Establishing species boundaries and new taxa among fungi: recommendations to resolve taxonomic ambiguities. Mycosphere 7: 1669–1699. 10.5943/mycosphere/7/11/4 [DOI] [Google Scholar]
- Katoh K, Standley DM. (2013) MAFFT Multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution 30: 772–780. 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langeron M, Milochevitch S. (1930) Annls Parasitol. Humaine Comp 8: 489.
- Milochevich (1935) Annls Parasitol. Humaine Comp. 13: 559.
- Nannizzi A. (1934) Repertorio sistematico dei miceti dell’uomo e degli animali. 4: 1–557. [Google Scholar]
- Oorschot CAN van. (1980) A revision of Chrysosporium and allied genera. Studies in Mycology 20: 1–89. [Google Scholar]
- Orr GF, Kuehn HH. (1963) The genus Ctenomyces. Mycopathologia et Mycologia Applicata 21: 321–333. 10.1007/BF02052585 [DOI] [PubMed] [Google Scholar]
- Posada D, Crandall KA. (1998) Modeltest: testing the model of DNA substitution. Bioinformatics 14: 817–818. 10.1093/bioinformatics/14.9.817 [DOI] [PubMed] [Google Scholar]
- Shadzi S, Chadeganipour M, Alimoradi M. (2002) Isolation of keratinophilic fungi from elementary schools and public parks in Isfahan, Iran. Mycoses 45: 496–499. 10.1046/j.1439-0507.2002.00798.x [DOI] [PubMed] [Google Scholar]
- Sharma M, Sharma M. (2010) Incidence of dermatophytes and other keratinophilic fungi in the schools and college playground soils of Jaipur, India. African Journal of Microbiology Research 4: 2647–2654. [Google Scholar]
- Silvestro D, Michalak I. (2012) raxmlGUI: a graphical front-end for RAxML. Organisms Diversity & Evolution 12: 335–337. 10.1007/s13127-011-0056-0 [DOI] [Google Scholar]
- Stamatakis A, Alachiotis N. (2010) Time and memory efficient likelihood-based tree searches on phylogenomic alignments with missing data. Bioinformatics 26: i132–i139. 10.1093/bioinformatics/btq205 [DOI] [PMC free article] [PubMed]
- Swofford DL. (2002) PAUP* 4.0b10: phylogenetic analysis using parsimony (*and other methods). Sunderland, MA: Sinauer. 10.1111/j.0014-3820.2002.tb00191.x [DOI]
- Szathmáry S. (1960) Die Cleistothecium-Bildung von Ctenomycestrichophyticus-Trichophytongypseumvar.asteroides-auf sterilisiertem Heu. Mykosen 3: 77–83. 10.1111/j.1439-0507.1960.tb03275.x [DOI] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Molecular Biology and Evolution 30: 2725–2729. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaidya G, Lohman DJ, Meier R. (2011) SequenceMatrix: concatenation software for the fast assembly of multi-gene datasets with character set and codon information. Cladistics 27: 171–180. 10.1111/j.1096-0031.2010.00329.x [DOI] [PubMed] [Google Scholar]
- Vanbreuseghem R. (1952) Technique biologique pour l’isolement des dermatophytes du sol. Annales de la Société Belge de Medicine Tropicale 32: 175–178. [PubMed] [Google Scholar]
- van den Brink J, Samson RA, Hagen F, Boekhout T, de Vries RP. (2012) Phylogeny of the industrial relevant, thermophilic genera Myceliophthora and Corynascus. Fungal Diversity 52: 197–207. 10.1007/s13225-011-0107-z [DOI] [Google Scholar]
- White TJ, Bruns T, Lee SJ, Taylor JL. (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR protocols: a guide to methods and applications 18: 315–322. 10.1016/B978-0-12-372180-8.50042-1 [DOI] [Google Scholar]
- Wijayawardene NN, Hyde KD, Lumbsch T, Liu JK, Maharachchikumbura SSN, Ekanayaka AH, Tian Q, Phookamsak R. (2018) Outline of Ascomycota – 2017. Fungal Diversity 88: 167–263. 10.1007/s13225-017-0386-0 [DOI] [Google Scholar]
- Wijayawardene NN, Hyde KD, Rajeshkumar KC, Hawksworth DL, Madrid H, Kirk PM, Braun U, Singh RV, Crous PW, Kukwa M, Lücking R, Kurtzman CP, Yurkov A, Haelewaters D, Aptroot A, Lumbsch HT, Timdal E, Ertz D, Etayo J, Phillips AJL, Groenewald JZ, Papizadeh M, Selbmann L, Dayarathne MC, Weerakoon G, Jones EBG, Suetrong S, Tian Q, Castañeda-Ruiz RF, Bahkali AH, Pang KL, Tanaka K, Dai DQ, Sakayaroj J, Hujslová M, Lombard L, Shenoy BD, Suija A, Maharachchikumbura SSN, Thambugala KM, Wanasinghe DN, Sharma BO, Gaikwad S, Pandit G, Zucconi L, Onofri S, Egidi E, Raja HA, Kodsueb R, Cáceres MES, Pérez-Ortega S, Fiuza PO, Monteiro JS, Vasilyeva LN, Shivas RG, Prieto M, Wedin M, Olariaga I, Lateef AA, Agrawal Y, Fazeli SAS, Amoozegar MA, Zhao GZ, Pfliegler WP, Sharma G, Oset M, Abdel MA, Takamatsu S, Bensch K, De Silva NI, De Kesel A, Karunarathna A, Boonmee S, Pfister DH, Lu YZ, Luo ZL, Boonyuen N, Daranagama DA, Senanayake IC, Jayasiri SC, Samarakoon MC, Zeng XY, Doilom M, Quijada L, Rampadarath S, Heredia G, Dissanayake AJ, Jayawardana RS, Perera PH, Tang LZ, Phukhamsakda C, Hernández-Restrepo M, Ma XY, Tibpromma S, Gusmao LFP, Weerahewa D, Karunarathna SC. (2017) Notes for genera: Ascomycota. Fungal Diversity 86: 1–594. 10.1007/s13225-017-0386-0 [DOI] [Google Scholar]
- Zarrin M, Haghgoo R. (2011) Survey of keratinophilic fungi from soils in Ahvaz, Iran. Jundishapur Journal of Microbiology 4: 191–194. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.