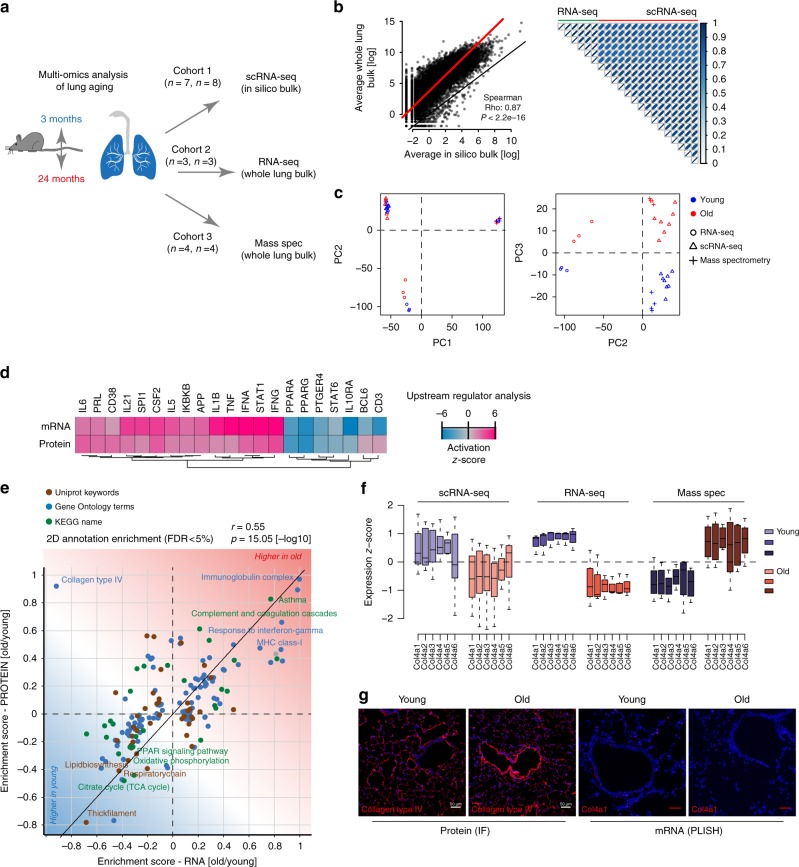
Fig. 3.
Multi-omic data integration uncovers uncoupled regulation of RNA and protein. a Experimental design—three independent cohorts of young and old mice were analyzed by single-cell RNA-sequencing (scRNA-seq), bulk RNA-seq, and mass spectrometry-driven proteomics respectively. b On the left, gene expression profiles from whole lung bulk samples (n = 6) and in silico bulk samples (n = 15) were averaged and plotted on X and Y axes, respectively. Red and black lines indicate linear model fit and the diagonal. On the right, correlation heatmap shows Pearson's correlation between all bulk and in silico bulk samples. c Normalized bulk (RNA-seq) and in silico bulk (scRNA-seq) data were merged with proteome data (mass spectrometry) and quantile normalized. The first two principal components show clustering by data modality. The third principal component separates young from old samples across all three data modalities. Blue and red colors indicate young and old samples, respectively. d Gene expression and protein abundance fold changes were used to predict upstream regulators that are known to drive gene expression responses similar to the ones experimentally observed. Upstream regulators could be cytokines or transcription factors. The color-coded activation z-score illustrates the prediction of increased or decreased activity upon aging. e The scatter plot shows the result of a two-dimensional annotation enrichment analysis based on fold changes in the transcriptome (x-axis) and proteome (y-axis), which resulted in a significant positive correlation of both datasets. Types of databases used for gene annotation are color coded as depicted in the legend. f Expression of collagen IV genes in the in silico bulk (scRNA-seq), bulk (RNA-seq), and proteome (mass spec) experiments, respectively. The box represents the interquartile range, the horizontal line in the box is the median, and the whiskers represent 1.5 times the interquartile range. g Immunofluorescence image of collagen type IV using confocal microscopy at ×25 magnification and proximity ligation in situ hybridization (PLISH) staining of Col4a1 mRNA. Note the increased fluorescence intensity of the protein around vessels in old mice along with the decreased mRNA expression (scale bar: 50 µm)