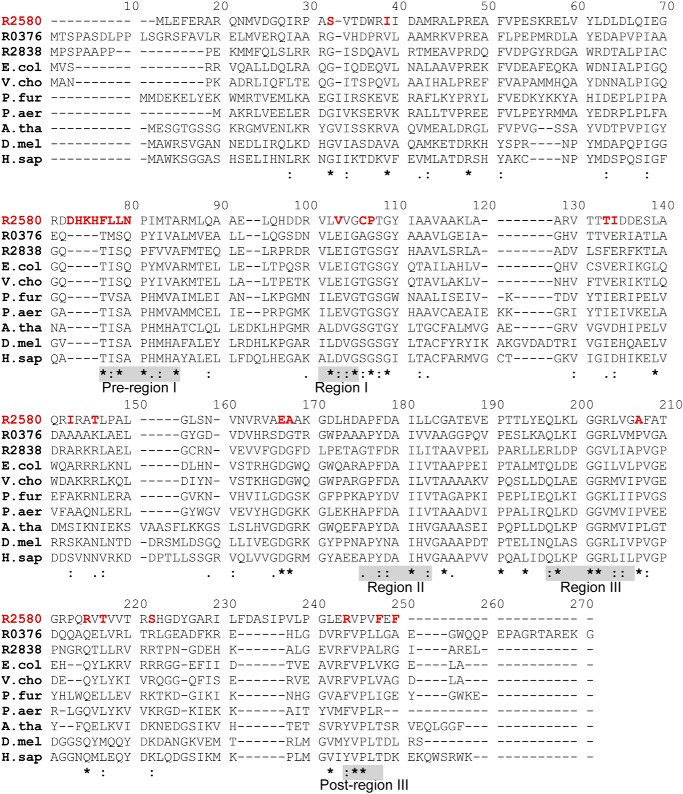
Figure 3.
Sequence divergence of RPA2580 from PIMT proteins. The alignment of the following 10 PIMT proteins is shown: R2580, RPA2580 (Q6N6N4); R0376, RPA0376 (Q6NCU3); R2838, RPA2838 (Q6N5Y0); E.col, PIMT in E. coli (P0A7A5); V.cho, PIMT in Vibrio cholera (Q9KUI8); P.fur, PIMT in P. furiosus (Q8TZR3); P.aer, PIMT in Pyrobaculum aerophilum (Q8ZYN0); A.tha, PIMT isoform 3 in Arabidopsis thaliana (Q64J17–3); D.mel, PIMT in D. melanogaster (Q27869); and H.sap, PIMT in Homo sapiens (P22061). Pre-region I and post-region III, which are characteristic of PIMTs, and regions I, II, and III, which are characteristic of this family of methyltransferases, are highlighted by shaded boxes (28). RPA2580 residues divergent from those in other PIMTs are highlighted with red type. Conserved PIMT amino acids that may not be conserved in RPA2580 are labeled as follows: asterisk, identical; colon, highly similar; period, similar.