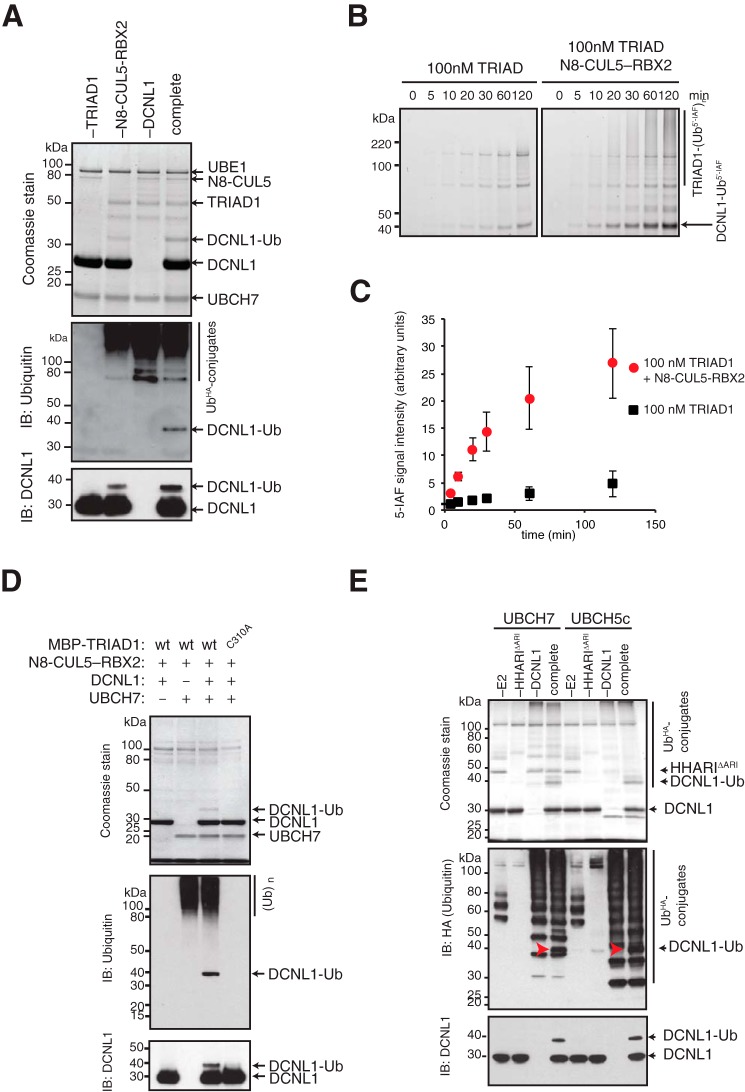
Figure 2.
TRIAD1 and HHARI monoubiquitylate DCNL1 in vitro. A, reconstitution of DCNL1 monoubiquitylation with purified recombinant TRIAD1, neddylated CUL5–RBX2 (N8–CUL5–RBX2), and UBCH7. Products of complete and drop-out (−) reactions were separated on SDS-PAGE and detected by Coomassie stain as well as immunoblot analysis as indicated. B, quantitative ubiquitylation assay with fluorescein-labeled ubiquitin (Ub5′-IAF) in the absence or presence of N8–CUL5–RBX2. Samples were taken at indicated time points and resolved on SDS-PAGE and scanned at 520 nm to visualize reaction products. C, quantitation of DCNL1-Ub signal from B using ImageJ software. Standard error of the mean is given from two independent replicates. D, DCNL1 monoubiquitylation with purified recombinant WT MBP–TRIAD1 or catalytically dead mutant MBP–TRIAD1 (C310A) with drop-out (−) UBCH7 and DCNL1 controls. Reaction products were separated on SDS-PAGE and detected by Coomassie stain as well as immunoblot analysis as indicated. E, DCNL1 monoubiquitylation reactions with HHARI that lacks the autoinhibitory Ariadne domain HHARI (ΔARI) using UBCH7 or UBCH5c as E2 enzymes including drop-out (−) E2, E3, and DCNL1 controls. Reaction products were separated on SDS-PAGE and detected by Coomassie stain and immunoblot analysis as indicated.