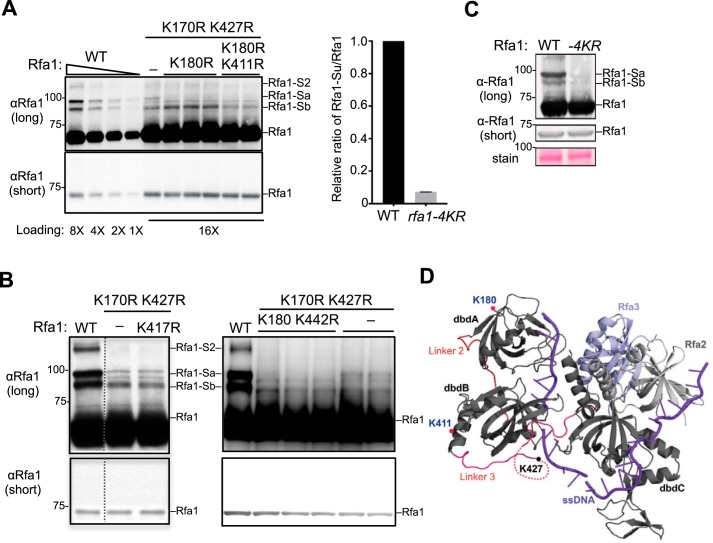
Figure 2.
Identification of two additional Rfa1 sumoylation sites. A, Mutating lysines 180 and 411 to arginine further reduces Rfa1 sumoylation. Adding the K180R mutation to rfa1-K170, 427R reduced the intensity of the Rfa1-Sa band. Further addition of the K411R mutation to rfa1-K170, 180, 427R (to give rfa1–4KR) reduced the intensity of the Rfa1-Sb band. The relative ratios of sumoylated Rfa1 forms to the unmodified Rfa1 form for WT and rfa1–4KR strains were quantified from two biological duplicates shown in the graph on the right. As Rfa1 sumoylation levels were low in rfa1-K170, 427R background, better visualization of this modification was achieved by using mec1Δ sml1Δ cells treated with 0.02% MMS, which are known to increase Rfa1 sumoylation. Note: For this experiment, rfa1Δ cells were supplemented with CEN-based plasmids expressing either WT or mutant Rfa1 driven by Rfa1 endogenous promoter. B, mutating lysine 417 and 442 to arginine does not affect Rfa1 sumoylation. Experiments were done as in panel A. Adding the K417R mutation to rfa1-K170, 427R had no effect on the intensity of all Rfa1 sumoylation bands. Similarly, adding the K442R mutation to rfa1-K170, 180, 427R did not further affect Rfa1 sumoylation. The data presented here are from the same immunoblot and dashed line indicates removal of superfluous lanes. C, rfa1–4KR as the only copy of Rfa1 in its endogenous locus greatly reduced Rfa1 sumoylation when cells were treated with 0.2% MMS for 2 h. rfa1–4KR did not affect Rfa1 protein levels as shown in the short exposure of the immunoblots. D, crystal structure of Ustilago maydis RPA in complex with ssDNA (purple) (25). Rfa1 is represented by its dbd A, B, and C. The locations of three of the four sumoylation sites are indicated. The K170 residue of Rfa1 was not included in the crystal structure.