Figure 6.
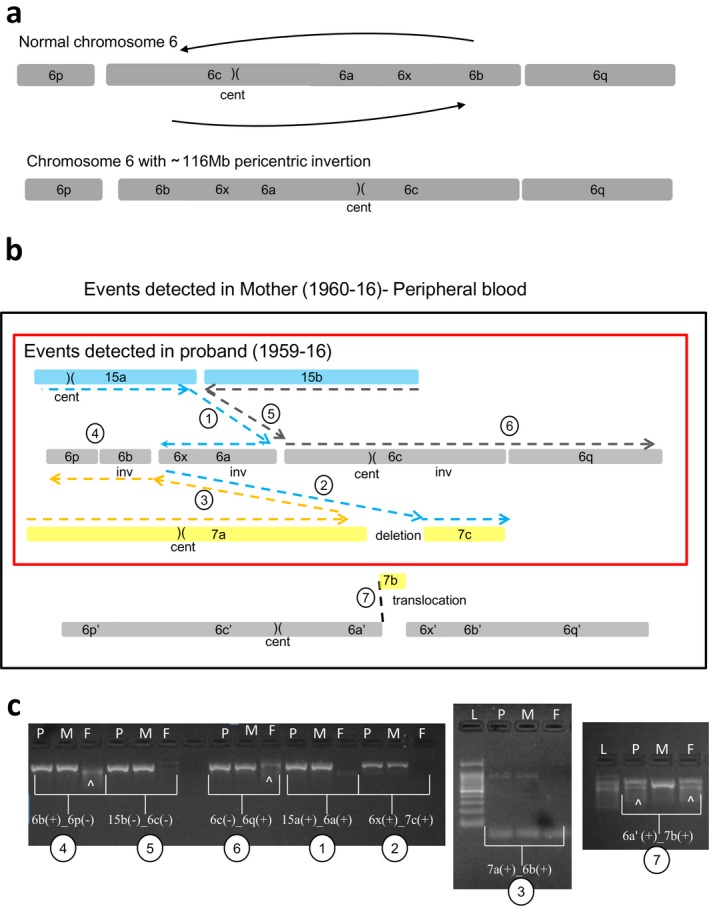
Case 3: Schematic view of t(6;7;15) chromothripsis of mother (1960‐16) and proband (1959‐16) (fragment size not in scale). (a) Shattering of chromosome 6 as a result of t(6;7;15) produced the fragments 6p, 6b, 6x, 6a, 6c, and 6q, altogether involving a 116 Mb pericentric inversion of chromosome 6 (cent: centromere). (b) Rearrangements of t(6;7;15) in mother's peripheral blood DNA involving mother‐specific 6a′‐7b breakpoint junction. Reconstruction of derivative chromosomes is illustrated with dashed lines, blue for derivative chromosome 15, gray for derivative chromosome 6, and yellow for derivative chromosome 7. Each fusion junctions are numbered from 1 to 7. The rearrangement detected in the proband is shown in a red rectangle. (c) Breakpoint‐specific PCR for the chromothripsis t(6;7;15) performed on proband (1959‐16), mother (1960‐16), and father (1961‐16). Case‐specific fusion junctions 6b(+)_6p(−) (number 4), 15b(−)_6c(−) (number 5), 6c(−)_6q(+) (number 6), 15a(+)_6a(+) (number 1), 6x(+)_7c(+) (number 2), and 7a(+)_6b(+) (number 3) are verified by the amplification of around 800 bp target size both in the mother and proband, but not in the father as expected. Fusion junction 6a′(+)_7b(+) (number 7) was verified only in the mother's DNA. The data proved that proband inherited all the breakpoints of t(6;7;15) except the fusion junction 6a′(+)_7b(+) (c). P: proband, M: mother, F: father, L: Ladder (GelPilot 100 bp plus ladder; Qiagen), (^): signifies nonspecific amplification
