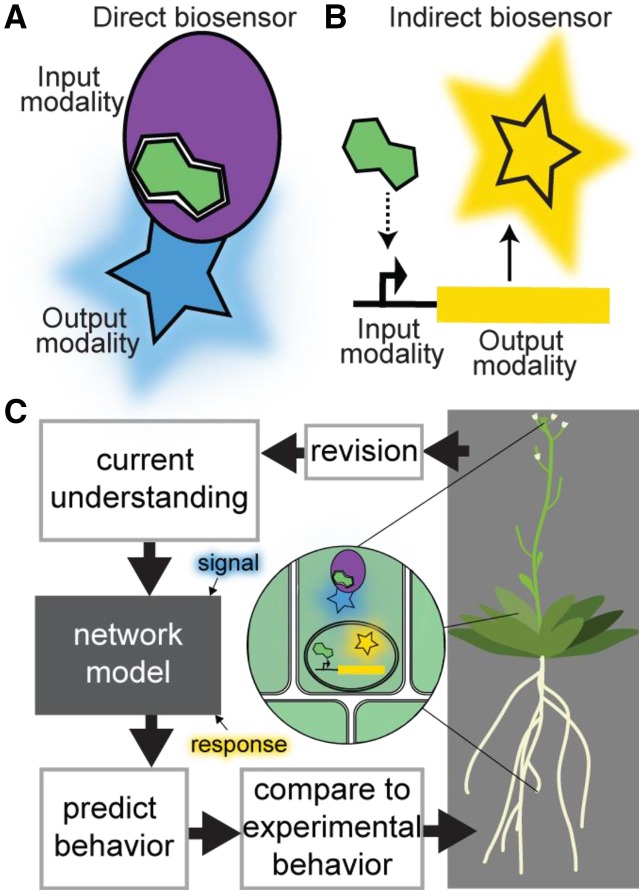
Figure 1.
Biosensors link detection of an analyte (such as a signaling molecule) by an input modality to a quantifiable change in an output modality. A, Schematic of a direct biosensor exemplified by a signaling molecule (green) binding protein as the input modality (purple oval) with a fluorescent protein output modality (blue star). This biosensor directly measures the “signal,” i.e. concentration of the signaling molecule. B, Schematic of an indirect biosensor exemplified by a signaling molecule responsive promoter of unknown mechanism (dotted arrow) driving expression of a fluorescent protein output modality (yellow star). This biosensor provides a measure of the response of this signaling pathway. C, Using biosensors to measure both the signal and response of a developmental signaling network along with plant phenotype leads to iterative improvement of the developmental network model and our understanding of plant development. Improved understanding of auxin signaling dynamics—realized by multiple biosensors and means of functional quantification—has facilitated rational tuning of plant architecture (Guseman et al., 2015; Je et al., 2016; Wright et al., 2017; Khakhar et al., 2018; Shibata et al., 2018). Newly developed biosensors (Liao et al., 2015; Rizza et al., 2017; Wu et al., 2018), paired with functional and phenotypic quantification of development, will help crack the code underlying developmental signaling and allow rational breeding and engineering of next-generation crops.