Figure 1.
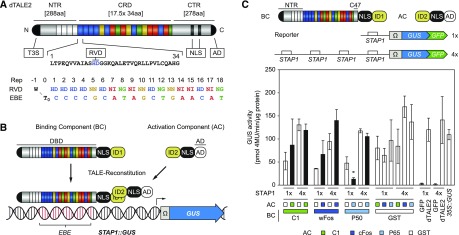
Design of the split-TALE and preliminary tests. A, Schematic representation of dTALE2 (NTR; central repeat region [CRD]; CTR; type 3 secretion signal [T3S]; RVD; NLS; AD; repeat [Rep]; EBE; invariant thymine flanking the 5` end of the RVD-defined target sequence [T0]. B, Schematic representation of the split-TALE system (BC; AC; DBD; ID; STAP). C, GUS-assay after Agrobacterium-mediated transient expression of indicated split-TALE components (BC, AC) and the reporter construct (1×STAP-Ω-GUS-GFP, 4×STAP-Ω-GUS-GFP) in N. benthamiana leaves. The BC contains the full-length NTR and 47 amino acids of the CTR. Color codes indicate corresponding IDs (C1, green; wFos/cFos, dark blue; P50/P65, blue; GST, white). White bars, except for the positive controls on the right side (dTALE2 and 35S::GUS), are for assays where only one component of the sTALE with an ID, either BC or AC, was expressed. In that case, the other component was fused to GST, which should not interact with the ID, and therefore served as a negative control. Black bars are for assays where both functional components were expressed. The identity of the expressed component is indicated below the graph by green boxes or squares. Free GFP (35S::GFP) and dTALE2 were used to monitor background activity of the STAPs (1x or 4x) and TALE-mediated transcriptional induction of the reporter, respectively. 35S::GUS serves as a positive control. Error bars represent sd (sd) of three biological replicates. Asterisks indicate a significant difference in activity of the same BC tested with control AC (ID = GST; Student’s t test; *P-value ≤ 0.05).