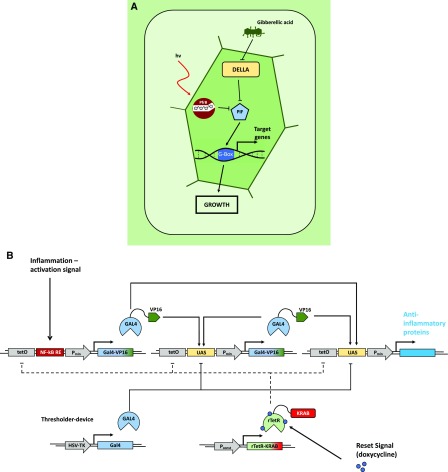
Figure 7.
Natural and engineered open-loop regulatory circuits. A, GA3-induced degradation of DELLA proteins suppresses the repression of PHYTOCHROME INTERACING FACTORs (PIFs). The PIFs subsequently bind to G-box cis regulatory elements in the promotors of response genes, promoting growth responses. In parallel, transcription of PIFs is inhibited by the red light-induced active conformer of phytochrome B, modulating the growth promotion in response to the light conditions. (Adapted from Havko et al., 2016.) B, Schematic overview of a synthetic device for detection of inflammation signals in mammalian systems. Detection of inflammatory signals through the NF-kB-responsive element of the sensor module leads to expression of the transcriptional regulator GAL4 fused to the VP16 transactivation domain (GAL4-VP16). GAL4-VP16 subsequently binds to the UAS motif in the amplifier and effector modules, increasing the abundance of GAL4-VP16 through a self-activating positive feedback loop from the amplifier module. This triggers production of anti-inflammatory proteins via the effector module. Additionally, the system is equipped with a thresholder device, constitutively expressing GAL4 lacking the transactivation domain. GAL4 competes for binding the UAS motifs with the activating GAL4-VP16, thereby restricting the initiation of the expression of the therapeutic output. A fifth module constitutively expresses the doxycycline-inducible reversed tetracycline repressor protein (rTetR) fused to the inhibitory KRAB domain. Exogenous application of doxycycline inhibits the activation of the sensor, amplifier, and effector modules by binding to their upstream tetO motifs, thus deactivating the system. (Adapted from Smole et al., 2017.)