The MoChlo kit is a modular tool for rapid assembly of chloroplast transformation vectors, designed to accelerate metabolic engineering in plastids.
Abstract
Plant synthetic biology is a rapidly evolving field with new tools constantly emerging to drive innovation. Of particular interest is the application of synthetic biology to chloroplast biotechnology to generate plants capable of producing new metabolites, vaccines, biofuels, and high-value chemicals. Progress made in the assembly of large DNA molecules, composing multiple transcriptional units, has significantly aided in the ability to rapidly construct novel vectors for genetic engineering. In particular, Golden Gate assembly has provided a facile molecular tool for standardized assembly of synthetic genetic elements into larger DNA constructs. In this work, a complete modular chloroplast cloning system, MoChlo, was developed and validated for fast and flexible chloroplast engineering in plants. A library of 128 standardized chloroplast-specific parts (47 promoters, 38 5ʹ untranslated regions [5ʹUTRs], nine promoter:5ʹUTR fusions, 10 3ʹUTRs, 14 genes of interest, and 10 chloroplast-specific destination vectors) were mined from the literature and modified for use in MoChlo assembly, along with chloroplast-specific destination vectors. The strategy was validated by assembling synthetic operons of various sizes and determining the efficiency of assembly. This method was successfully used to generate chloroplast transformation vectors containing up to seven transcriptional units in a single vector (∼10.6-kb synthetic operon). To enable researchers with limited resources to engage in chloroplast biotechnology, and to accelerate progress in the field, the entire kit, as described, is available through Addgene at minimal cost. Thus, the MoChlo kit represents a valuable tool for fast and flexible design of heterologous metabolic pathways for plastid metabolic engineering.
The plastids of land plants represent an underutilized resource with regard to metabolic engineering of heterologous pathways (Maliga and Bock, 2011; Bock, 2013; Jin and Daniell, 2015; Schindel et al., 2018). Plastids have been successfully engineered for expression of valuable carotenoids (Wurbs et al., 2007; Apel and Bock, 2009) and terpenoids (Pasoreck et al., 2016) as well as for the production of numerous biopharmaceuticals (Davoodi-Semiromi et al., 2010; Boyhan and Daniell, 2011; Lakshmi et al., 2013). Nonetheless, there are far fewer instances in which the plastid chassis has been engineered for crop improvement traits. Certainly, a more controlled metabolic engineering approach would facilitate a fundamental change in crop improvement (Schindel et al., 2018). One such example, using plastid engineering, is the metabolic engineering of a complete carbon-concentrating mechanism from cyanobacterium into plastids for improving photosynthesis (Lin et al., 2014a, 2014b; Occhialini et al., 2016; Long et al., 2018). Since plastid genomes are relatively conserved among land plants, improving photosynthesis in one species via plastids should be translatable to other species (McNeal et al., 2007).
There are several advantages to engineering plastid genomes, as opposed to nuclear ones, for metabolic engineering in plants. Plastids have small genomes (120–217 kb) present in multiple copies (e.g. ∼10,000 plastome copies per cell in tobacco [Nicotiana tabacum]), lack gene-silencing machinery, and are capable of extremely high levels of foreign protein production (10%–70% total soluble protein). Also, transgene insertion is achieved through homologous recombination, providing a known landing site for insertion (Kuroda and Maliga, 2001a; Maliga, 2004; Shaver et al., 2006; Verma and Daniell, 2007; Verma et al., 2008; Oey et al., 2009a; Schelkunov et al., 2015). Furthermore, since chloroplasts are prokaryotic in origin, the transcription and translation machinery functions similar to prokaryotes, allowing for coordinated gene expression in operons (Bock, 2013; Lu et al., 2013). For metabolic engineering, the ability to coordinate the expression of multiple genes is essential for controlling complex multigene pathways. Unfortunately, while the regulatory machinery of plastids is prokaryotic in origin, the governing rules/functional motifs used for gene expression are not nearly as well characterized as their bacterial counterparts. Thus, while numerous logic circuits and tunable expression systems have been developed in bacterial systems, few exist for plastids (Verhounig et al., 2010; Emadpour et al., 2015). Instead, most complex pathways expressed in plastids use a single promoter to drive equivalent expression of all transgenes in the pathway (Lu et al., 2013; Fuentes et al., 2016). However, excess production of heterologous protein, even green fluorescent protein (GFP), has had demonstrable negative effects on chloroplast metabolic function and the overall phenotype of the resulting plants (Oey et al., 2009a). Advanced metabolic engineering in plastids requires an understanding of how to tune the stoichiometry of multiple genes within a heterologous pathway to achieve the desired product (Bock, 2013, 2015; Schindel et al., 2018).
Typically, heterologous genes engineered into plastids are placed under the control of endogenous regulatory sequences (Tregoning et al., 2003; Lutz et al., 2007; Zhou et al., 2008; Oey et al., 2009b). For effective chloroplast metabolic engineering with tunable expression, the following components must be considered: promoters, 5ʹ untranslated regions (5ʹUTRs), and 3ʹUTRs. Precise regulation of gene expression in plastids could be controlled by varying these components to achieve the desired expression level for each transgene or operon. While combinatorial and permutational analyses of these components in a bacterial chassis, such as Escherichia coli, are routinely conducted, plastids represent a more challenging system because the plastid toolkit is much less developed than that for bacteria. In addition, compared with nuclear engineering in plants, few labs currently conduct plastid engineering.
In this article, we describe a plastid molecular cloning kit, MoChlo, for plastid genetic engineering in plants. The MoChlo kit builds on previously developed modular cloning kits (Sarrion-Perdigones et al., 2011; Weber et al., 2011; Lampropoulos et al., 2013; Engler et al., 2014; Vafaee et al., 2014). Our goals are to lower the barrier of entry into plastid engineering, to enable troubleshooting for transgene expression, and to provide more flexibility for plastid metabolic engineering. Our hope is that a standardized assembly kit composed of a library of plastid regulatory elements will accelerate research into metabolic engineering in plastids and benefit the research community as a whole.
RESULTS
MoChlo Kit Design
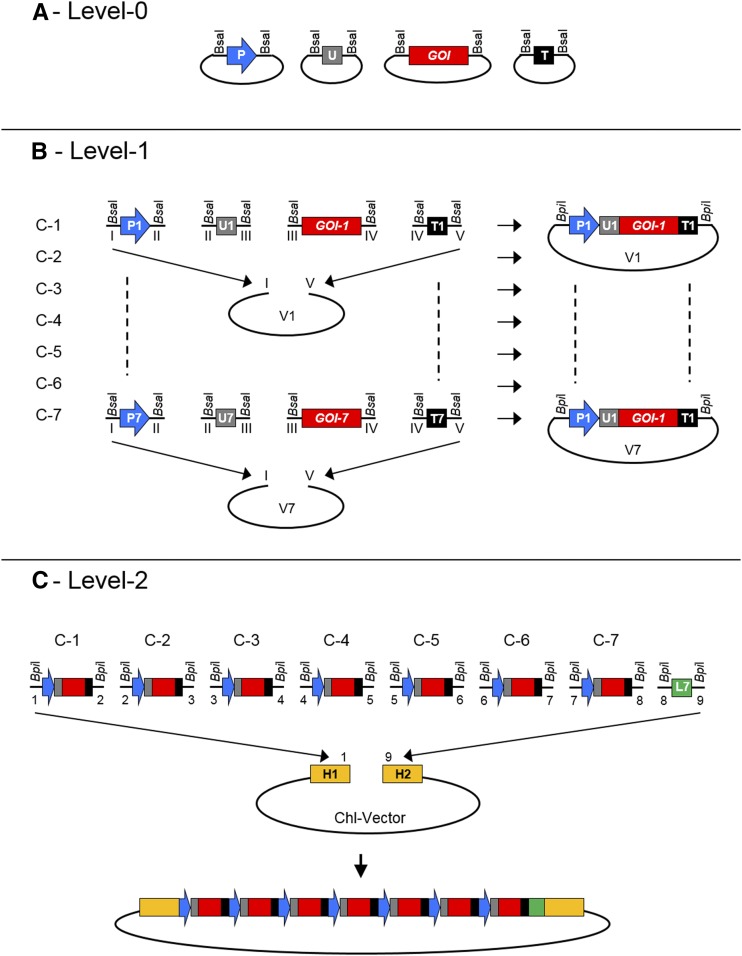
The nomenclature and strategy used for the various levels and components of the MoChlo system were adopted from previous plant MoClo kits (Fig. 1; Supplemental Fig. S1; Weber et al., 2011; Engler et al., 2014). All individual parts (Level-0) mined from the literature were domesticated using the criteria described in “Materials and Methods.” The domesticated MoChlo parts library comprises 47 chloroplast promoters, 38 5ʹUTRs, nine fused chloroplast promoter::5ʹUTRs, and 10 terminators/3ʹUTRs. In addition, the library includes 14 coding genes including selectable markers (e.g. aadA) and reporter genes (e.g. mEmerald GFP). A complete list of all Level-0 modules provided in the MoChlo kit, including references to where the sequence was mined, is shown in Table 1 and Supplemental Table S1. All 14 Level-1 destination vectors (seven in forward and seven in reverse orientation) were obtained directly from a previous MoClo kit (Engler et al., 2014; Supplemental Table S2). With the current components of the MoChlo kit, it is possible to assemble up to seven Level-1 constructs into a single Level-2 construct (Fig. 1; Supplemental Fig. S2). In addition, a set of seven end linkers compatible with the 3ʹ overhang of each position of the final operon provides the option to integrate a variable number of cassettes into Level-2 vectors (Supplemental Fig. S2; Supplemental Table S3). The final components of the MoChlo system are the Level-2 chloroplast-specific destination vectors. The Level-2 destination vectors in the MoChlo system were developed for three plant species (tobacco, maize [Zea mays], and potato [Solanum tuberosum]) with homologous arms designed for recombination into four different integration sites of the plastome (trnI-trnA, trnG-trnfM, trnV-3ʹrps12, and atpB-accD; Supplemental Table S3). Thus, in the current version of the MoChlo kit, researchers would be able to directly assemble chloroplast transformation cassettes for multiple plant species utilizing a variety of regulatory elements as well as selectable marker genes.
Figure 1.
Schematic representation of Golden Gate assembly using the MoChlo kit. A, Four vectors containing different Level-0 modules flanked by BsaI sites. P, Promoter (blue); U, 5ʹUTR (gray); GOI, gene of interest (red); T, terminator/3ʹUTR (black). B, Level-1 Golden Gate assembly. The BsaI restriction digestion releases Level-0 parts (P1–P7, U1–U7, GOI1–GOI7, and T1–T7) equipped with 5ʹ and 3ʹ overhangs (I–V) used for directional cloning. The 5ʹ and 3ʹ overhangs (I and V, respectively) of P-U-GOI-T cassettes (C1–C7) are compatible with Level-1 vectors (V1–V7). The Level-1 assembly results in seven Level-1 vectors containing seven cassettes (C1–C7) flanked by BpiI sites. C, Level-2 Golden Gate assembly. The BpiI restriction digestion of Level-1 vectors releases cassettes (C1–C7) equipped with 5ʹ and 3ʹ overhangs (1–8) able to provide directional cloning from C1 to C7. An end linker (L7; green) equipped with compatible 5ʹ and 3ʹ overhangs (8 and 9, respectively) allows integration of the Level-2 operon in between the homologous sequences (H1 and H2) of a chloroplast transformation vector (Chl-Vector; destination vector Level-2).
Table 1. Level-0 parts provided in the MoChlo kit.
A detailed description of all Level-0 parts, including nucleotide sequences, is included in Supplemental Table S1. (–), No data; u, unpublished; NEP, nuclear encoded polymerase; PEP, plastid encoded polymerase; DB, downstream box; Ec, Escherichia coli; TYMV, turnip yellow mosaic virus; BMV, brome mosaic virus; AMV, Alfalfa mosaic virus; TMV, tobacco mosaic virus; SL, stem-loop; codon opt., plastid codon optimized.
| Promoters | 5′UTR | ||
|---|---|---|---|
| PaccD (−82 to +23; Hirata et al., 2004) | Prrn16 (Allison et al., 1996) | atpB 5′UTR (Hirose and Sugiura, 2004) | rbcL 5′UTR (full; Yukawa et al., 2007) |
| PaccD (functional; Hirata et al., 2004) | Prrn (tobacco; Suzuki et al., 2003) | atpB 5′UTR +10 (functional; Hirose and Sugiura, 2004) | rbcL 5′UTR (processed; Yukawa et al., 2007) |
| PatpB (NEP functional PEP disabled; Xie and Allison, 2002) | Synthetic APrrn1u | atpB 5′UTR +10 processed −90 (functional; Yukawa et al., 2007) | rbcL 5′UTR + 14 AA (Kuroda and Maliga, 2001b) |
| PatpB (NEP; Xie and Allison, 2002) | Synthetic APrrn2u | atpB 5′UTR TCR processed −90 (functional; Kuroda and Maliga, 2001b) | Synthetic RBS (Kuroda and Maliga, 2001a) |
| PatpB (PEP and NEP functional; Xie and Allison, 2002) | Synthetic APrrn3u | atpH 5′UTR (Yukawa et al., 2007) | T7G10 leader (Kuroda and Maliga, 2001a) |
| PatpB (PEP functional NEP disabled; Xie and Allison, 2002) | Synthetic APrrn4u | atpH 5′UTR processed −45 (Yukawa et al., 2007) | T7G10 leader + Ec DB (Kuroda and Maliga, 2001a) |
| PatpB (PEP; Xie and Allison, 2002) | Synthetic APrrn5u | atpI 5′UTR (Baecker et al., 2009) | rrn16 5′UTR (native; Allison et al., 1996) |
| PatpH (Miyagi et al., 1998) | Synthetic APrrn6u | psbN 5′UTR (Kuroda and Sugiura, 2014) | rrn16 5′UTR (+37; Allison et al., 1996) |
| PatpI (NEP; Miyagi et al., 1998) | Synthetic APrrn7u | psbN 5′UTR processed −39 (Kuroda and Sugiura, 2014) | psbA 5′UTR (Staub and Maliga, 1994) |
| PatpI (PEP hypothetical; Miyagi et al., 1998) | Synthetic APrrn8u | psbB 5′UTR (Yukawa et al., 2007) | Synthetic RBS (Allison and Maliga, 1995) |
| PclpP (Sriraman et al., 1998) | Synthetic APrrn9u | psbB 5′UTR processed −54 (Yukawa et al., 2007) | psbC 5′UTR (Kuroda et al., 2007) |
| PclpP (PEP; Sriraman et al., 1998) | Synthetic APrrn10u | ndhD 5′UTR (Hirose and Sugiura, 1996) | accD 5′UTR (Caroca et al., 2013) |
| IEE (Zhou et al., 2007) | Synthetic BPrrn12u | rps2 5′UTR (Plader and Sugiura, 2003) | clpP 5′UTR (Kuroda and Maliga, 2002) |
| Prrn (−64 to +17; Suzuki et al., 2003) | Synthetic BPrrn13u | rps2 5′UTR mAAGA (Plader and Sugiura, 2003) | – |
| mPrrn (5-fold decrease transcription; Suzuki et al., 2003) | Synthetic BPrrn14u | rps2 5′UTR mCCUC (Plader and Sugiura, 2003) | – |
| PpsbA (Staub and Maliga, 1994) | Synthetic BPrrn15u | rps2 5′UTR mAAGA (−20 to −10 deletion; Plader and Sugiura, 2003) | – |
| PpsbA (−42 to +9; Hayashi et al., 2003) | Synthetic BPrrn16u | rps2 5′UTR mCCUC (−20 to −10 deletion; Plader and Sugiura, 2003) | – |
| PpsbB (Tanaka et al., 1987) | Synthetic BPrrn17u | psbH 5′UTR + start codon + 1 AA (Hammani et al., 2012) | – |
| PpsbD leader (Allison and Maliga, 1995) | Synthetic BPrrn18u | psbA 5′UTR (SNM; Zou et al., 2003) | – |
| PpsbD-PGTbx (Allison and Maliga, 1995) | Synthetic BPrrn19u | psbA 5′UTR (SEM; Zou et al., 2003) | – |
| PpsbD (Allison and Maliga, 1995) | Synthetic BPrrn20u | psbA 5′UTR (delta SL; Zou et al., 2003) | – |
| PrbcL (Shiina et al., 1998) | Synthetic BPrrn21u | psbA 5′UTR (delta 5′SL; Zou et al., 2003) | – |
| PrbcL (core −3 to +9; Shiina et al., 1998) | – | psbA-rbcL chimeric 5′UTR (R delta 5′SL; Zou et al., 2003) | – |
| PrbcL (−110 to +9; Shiina et al., 1998) | – | psbA-rbcL chimeric 5′UTR (R′SLWT; Zou et al., 2003) | – |
| PrpoB (Liere and Maliga, 1999) | – | rps14 5′UTR (Hirose et al., 1998) | – |
| Promoter::5′UTR | GOI | Integration Sites | 3′UTR |
|---|---|---|---|
| Maize PclpP::5′UTR (Zhang et al., 2012) | mEmerald (Cubitt et al., 1999) | Tobacco trnI/trnA (Daniell et al., 1998) | psbA 3′UTR (Eibl et al., 1999) |
| Maize PclpP::Gg10 leader (Zhang et al., 2012) | pporRFP (Alieva et al., 2008) | Tobacco trnG/trnfM (Bock and Maliga, 1995) | rbcL 3′UTR (Eibl et al., 1999) |
| PaccD::5′UTR (Zhang et al., 2012) | aadA (Svab and Maliga, 1993) | Tobacco trnV/3′rps12 (Staub and Maliga, 1992) | petD 3′UTR (Tangphatsornruang et al., 2011) |
| mPrrn::RBS (3.9%; Suzuki et al., 2003) | aphA6 (Huang et al., 2002) | Tobacco atpB/accD (Lin et al., 2014b) | rpoA 3′UTR (Tangphatsornruang et al., 2011) |
| mPrrn::RBS (20%; Suzuki et al., 2003) | nptII (Carrer et al., 1993) | Maize trnI/trnA (Daniell et al., 1998) | E. coli rrnB 3′UTR (Tangphatsornruang et al., 2011) |
| mPrrn::RBS (46%; Suzuki et al., 2003) | Myc aadA-GFP (Lutz et al., 2007) | Maize trnV/3′rps12 (Staub and Maliga, 1992) | psaC 3′UTR (Hirose and Sugiura, 1997) |
| mPrrn::RBS (60%; Suzuki et al., 2003) | FLARE16-S aadA-GFP (Khan and Maliga, 1999) | Potato trnI/trnA (Daniell et al., 1998) | TMV 3′UTR (Kawamura-Nagaya et al., 2014) |
| mPrrn::RBS (61%; Suzuki et al., 2003) | aac6-aph2 (Tabatabaei et al., 2017) | Potato trnG/trnfM (Bock and Maliga, 1995) | TYMV 3′UTR (Ma et al., 2015) |
| Prrn::RBS (Daniell et al., 2005) | Lux-C codon opt. tobacco chloroplasts (Lee et al., 1991) | Potato trnV/3′rps12 (Staub and Maliga, 1992) | BMV 3′UTR (Bujarski et al., 1985) |
| – | Lux-D codon opt. tobacco chloroplasts (Lee et al., 1991) | Potato trnV/3′rps12 (Staub and Maliga, 1992) | ALMV 3′UTR (Trucco et al., 2014) |
| – | Lux-A codon opt. tobacco chloroplasts (Lee et al., 1991) | – | – |
| – | Lux-B codon opt. tobacco chloroplasts (Lee et al., 1991) | – | – |
| – | Lux-E codon opt. tobacco chloroplasts (Lee et al., 1991) | – | – |
| – | Lux-G codon opt. tobacco chloroplasts (Lee et al., 1991) | – | – |
Validation of the MoChlo Strategy for Assembly of Simple Synthetic Operons (Four or Fewer Transcriptional Units)
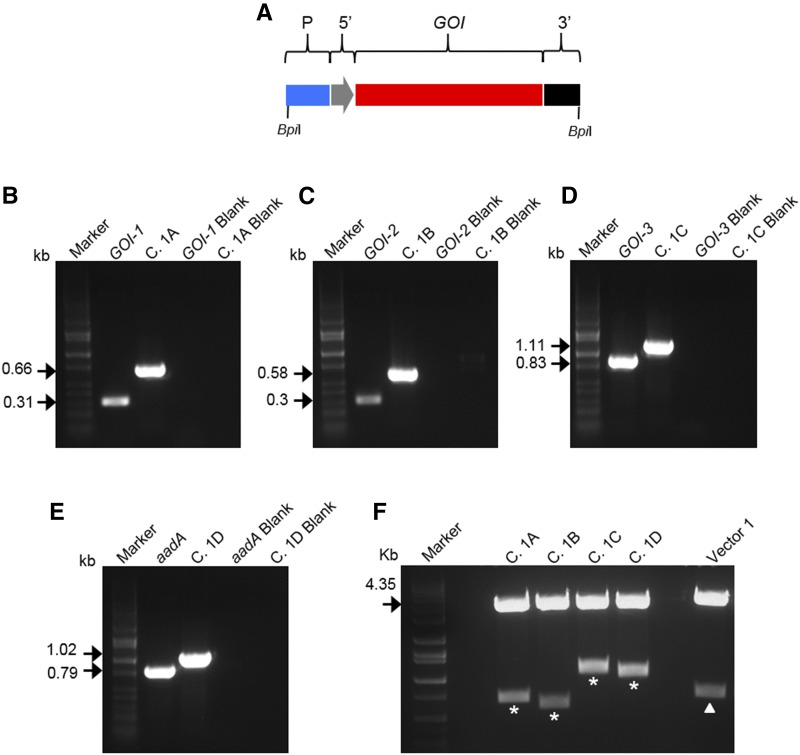
The first step in validating the MoChlo kit was assembling Level-1 constructs from the Level-0 parts (Fig. 2). Four Level-1 transcriptional units expressing GOI-1, GOI-2, GOI-3, or aadA were cloned into four Level-1 vectors for positions 1, 2, 3, and 4 of the operon, respectively. A description of regulatory sequences (Level-0 parts) used to assemble the aforementioned 1A, 1B, 1C, and 1D Level-1 cassette is shown in Table 2. The efficiency of assembly of the Level-1 constructs was 100%, as determined by blue-white screening, PCR, and restriction digestion (Fig. 2; Supplemental Fig. S3). Finally, the correct nucleotide sequence of all Level-1 constructs was verified by Sanger sequencing. Examples of another 14 Level-1 cassettes assembled using the MoChlo kit are shown in Supplemental Figure S4 and had the same efficiency of 100%.
Figure 2.
Level-1 cloning for a four-gene operon-1. A, Schematic representation of a Level-1 cassette: promoter (P; blue); 5ʹUTR (5ʹ; gray); GOIs (GOI1–GOI3 or aadA; red); terminator/3ʹUTR (3ʹ; black). BpiI sites flanking the Level-1 cassette are indicated. B to E, PCR to confirm Level-1 assembly. Specific primers for the internal gene (B, GOI-1; C, GOI-2; D, GOI-3; E, aadA) and the promoter/3ʹUTR of cassettes 1A, 1B, 1C, and 1D (B, C. 1A; C, C. 1B; D, C. 1C; E, C. 1D) have been used. DNA bands at the predicted molecular mass (kb) confirm correct Level-1 assembly of all cassettes. Negative controls (blanks) of PCR images B to E are indicated. F, BpiI restriction digestion of Level-1 vectors. The four Level-1 cassettes released after digestion are indicated with asterisks (size indicated in Table 1). The empty vector 1 (vector position 1 forward) is used as a positive control. The LacZα fragment (0.616 kb) release after digestion of vector 1 is indicated with the triangle. The molecular mass (kb) of the backbone vector is indicated with black arrows. DNA markers are shown in each image.
Table 2. Design of four different Level-1 cassettes.
| Name | Promoter | 5′UTR | GOI | 3′UTR | Size (kb) | Position (Forward) |
|---|---|---|---|---|---|---|
| Cassette 1A | PaccD | Synthetic RBS | GOI-1 | petD-3′UTR | 0.664 | 1 |
| Cassette 1B | IEE | Synthetic RBS | GOI-2 | petD-3′UTR | 0.583 | 2 |
| Cassette 1C | IEE | Synthetic RBS | GOI-3 | petD-3′UTR | 1.114 | 3 |
| Cassette 1D | PpsbA | Synthetic RBS | aadA | psbA-3′UTR | 1.019 | 4 |
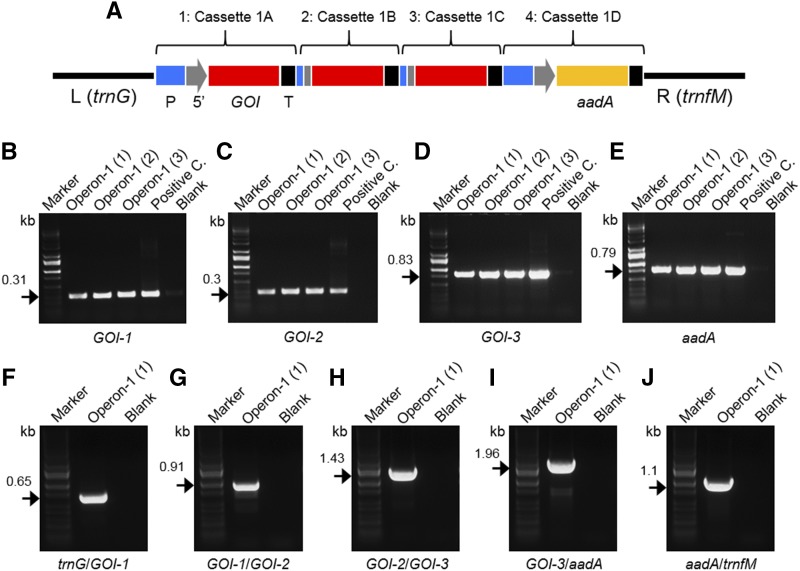
To validate Level-2 assembly, Level-2 operon-1 (∼3.5 kb) was assembled in a second assembly step using the four Level-1 cassettes (1A, 1B, 1C, and 1D) generated previously. For this assembly, a Level-2 destination vector equipped with homologous arms to trnG-trnfM from potato was used (Fig. 3). This operon was designed to represent a standard polycistronic operon (three GOI) along with a monocistronic selection cassette. Using BbsI to release all of the Level-1 cassettes for Level-2 assembly, the efficiency of Level-2 assembly, as determined by PCR, was 22.5% (Supplemental Fig. S5). However, when using the isoschizomer BpiI instead, GOI-1 was found to be present in all 40 colonies tested, with 37 of the colonies positive for all genes in the cassette (Supplemental Fig. S6). Thus, the efficiency of Level-2 assembly was 92.5% using BpiI instead of BbsI, as determined by colony PCR. With a simpler assembly combining only two Level-1 cassettes into Level-2, instead of the four described previously, there was no difference in efficiency between assembly using BbsI and BpiI, with both achieving 100% efficiency (Supplemental Fig. S7). Correct assembly of Level-2 operon-1 was further verified by PCR of the individual components (Fig. 3) as well as by PCR confirmation of the junctions between individual Level-1 constructs (trnG/GOI-1, GOI-1/GOI-2, GOI-2/GOI-3, GOI-3/aadA, and aadA/trnfM; Fig. 3). Examples of another four Level-2 operons assembled by the same four Level-1 cassettes are shown in Supplemental Figure S8.
Figure 3.
Level-2 cloning for operon-1. A, Schematic representation of Level-2 operon-1: promoter (P; blue); 5ʹUTR (5ʹ; gray); GOIs (GOI1–GOI3; red); gene for selection (aadA; yellow); terminator/3ʹUTR (3ʹ; black). Level-1 cassettes 1A, 1B, 1C, and 1D are assembled in positions 1, 2, 3, and 4 of operon-1, respectively. The operon-1 is cloned in a Level-2 vector between left (L; 1.17 kb) and right (R; 1.45 kb) arms homologous to the trnG/trnfM site of the potato plastome. B to E, PCR to confirm the presence of all genes of operon-1 (three purified vectors; 1–3). Specific primers for GOI-1 (B), GOI-2 (C), GOI-3 (D), and aadA (E) have been used. DNA bands at the predicted molecular mass (kb; black arrows) in both purified plasmid (operon-1) and positive controls (positive C.) confirm correct Level-2 assembly of operon-1. F to J, PCR to confirm the correct order of four genes in the operon-1. Pairs or primers specific for trnG/GOI-1 (F), GOI-1/GOI-2 (G), GOI-2/GOI-3 (H), GOI-3/aadA (I), and aadA/trnfM (J) have been used. DNA bands at the predicted molecular mass confirm the correct order (kb; black arrows). Negative controls (blanks) for PCR and DNA markers are shown in B to J.
Validation of MoChlo Strategy for Assembly of a Large Operon (Seven Transcriptional Units)
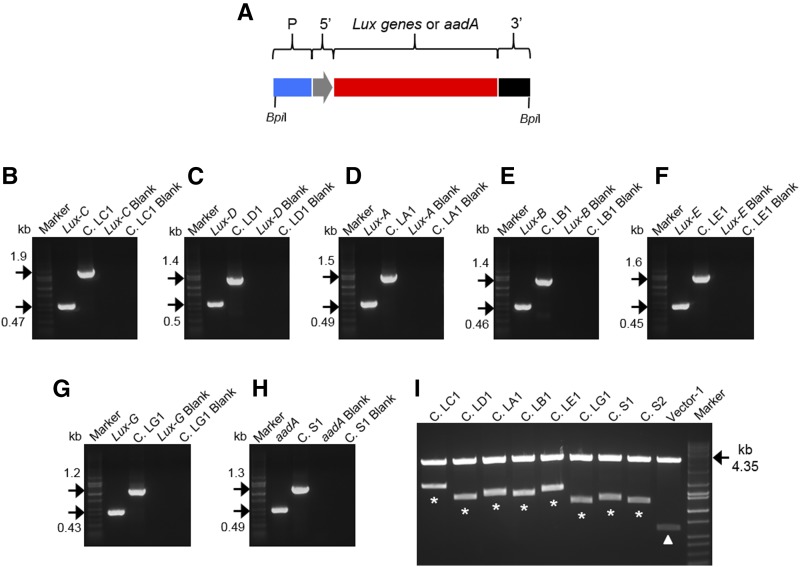
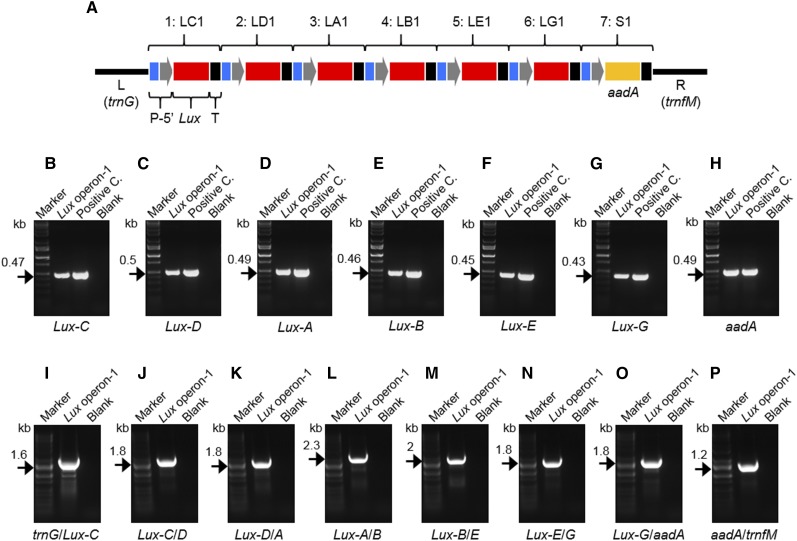
In the current iteration of the MoChlo kit, the maximum number of Level-1 constructs that can be combined into a single Level-2 cassette is seven. To test the efficiency of the maximum assembly, the six genes comprising the Lux operon (Lux-C, -D, -A, -B, -E, and -G) were domesticated to Level-0 parts and assembled into Level-1 constructs (LC1, LD1, LA1, LB1, LE1, and LG1) with similar efficiency to that described previously (Table 3; Fig. 4; Supplemental Fig. S9). The seventh position in the maximum Level-2 cassette was reserved for selection using either aadA or mEmerald (Cassette S1 and S2, respectively). After sequence verification of the Level-1 constructs, Level-2 assembly of the six Lux cassettes, along with Cassette S1, was conducted using the potato trnG-trnfM Level-2 chloroplast destination vector (Fig. 5). For this test case, the assembly was designed to express seven monocistronic transcriptional units (∼10.6 kb total), in which the seven genes are under the control of the same regulatory sequences (Table 3). After double selection with kanamycin/spectinomycin (nptII on Level-2 backbone and aadA within operon), it was determined that 28 of 40 colonies were PCR positive for the presence of Lux-C, with 26 of the 28 Lux-C-positive colonies positive for all genes of the operon (Supplemental Fig. S10). In this test case, the efficiency of assembly of seven Level-1 constructs into a single Level-2 construct was 65%. In the case of using BbsI instead of BpiI, the efficiency was substantially reduced to 1.25% (Supplemental Fig. S11). Correct assembly of the complete Level-2 operon was further validated using single-gene and junction PCR (Fig. 5). Thus, with the current design of the MoChlo kit, a single chloroplast transformation vector containing up to seven Level-1 cassettes can be assembled with high efficiency.
Table 3. Design of six Level-1 cassettes for the Lux operon and two Level-1 selection cassettes.
| Name | Promoter | 5′UTR | GOI | 3′UTR | Size (kb) | Position (Forward) |
|---|---|---|---|---|---|---|
| Cassette LC1 | Prrn | Prrn-5′UTR | Lux-C | rrnB-3′UTR | 1.919 | 1 |
| Cassette LD1 | Prrn | Prrn-5′UTR | Lux-D | rrnB-3′UTR | 1.430 | 2 |
| Cassette LA1 | Prrn | Prrn-5′UTR | Lux-A | rrnB-3′UTR | 1.547 | 3 |
| Cassette LB1 | Prrn | Prrn-5′UTR | Lux-B | rrnB-3′UTR | 1.463 | 4 |
| Cassette LE1 | Prrn | Prrn-5′UTR | Lux-E | rrnB-3′UTR | 1.604 | 5 |
| Cassette LG1 | Prrn | Prrn-5′UTR | Lux-G | rrnB-3′UTR | 1.187 | 6 |
| Cassette S1 | Prrn | Prrn-5′UTR | aadA | rrnB-3′UTR | 1.274 | 7 |
| Cassette S2 | Prrn | Prrn-5′UTR | mEmerald | rrnB-3′UTR | 1.202 | 7 |
Figure 4.
Level-1 cloning for the Lux operon for plastid engineering. A, Schematic representation of a Level-1 cassette: promoter (P; blue); 5ʹUTR (5ʹ; gray); Lux (C, D, A, B, E, or G) or aadA gene for selection (red); terminator/3ʹUTR (3ʹ; black). BpiI sites flanking the Level-1 cassette are indicated. B to H, PCR to confirm Level-1 assembly. Specific primers for Lux genes (B, Lux-C; C, Lux-D; D, Lux-A; E, Lux-B; F, Lux-E; G, Lux-G), the gene for selection (H; aadA), and the promoter/3ʹUTR of cassettes LC1, LD1, LA1, LB1, LE1, LG1, and S1 (B, C. LC1; C, C. LD1; D, C. LA1; E, C. LB1; F, C. LE1; G, C. LG1; H, C. S1) have been used. DNA bands at the predicted molecular mass (kb; black arrows) confirm correct Level-1 assembly of all cassettes. Negative controls (blanks) of PCR images B to H are indicated. I, BpiI restriction digestion of Level-1 vectors. Level-1 cassettes released after digestion are indicated with asterisks (cassette size indicated in Table 2). Empty vector 1 (position 1 forward) was used as a positive control. The LacZα fragment release after digestion of vector 1 is indicated with the arrowhead (0.616 kb). The molecular mass (kb) of the backbone vector is indicated with black arrows. DNA markers are shown in B to I.
Figure 5.
Level-2 cloning for the seven-gene Lux operon. A, Schematic representation of Level-2 assembly of Lux operon-1: promoter (P; blue); 5ʹUTR (5ʹ; gray); Lux (C, D, A, B, E, or G; red); gene for selection (aadA; yellow); terminator/3ʹUTR (3ʹ; black). Level-1 cassettes LC1, LD1, LA1, LB1, LE1, LG1, and S1 are assembled in operon positions 1, 2, 3, 4, 5, 6, and 7, respectively. The Lux operon-1 is cloned in Level-2 vector between left (L; 1.17 kb) and right (R; 1.45 kb) arms homologous to the trnG/trnfM site of the potato plastome. B to H, PCR of all genes of the Lux operon-1. Primers specific for Lux-C (B), Lux-D (C), Lux-A (D), Lux-B (E), Lux-E (F), Lux-G (G), and aadA (H) have been used. DNA bands at the predicted molecular mass (kb; black arrows) for both purified plasmid (Lux operon-1) and positive controls (positive C.) confirmed the presence of all genes. I to P, PCR to check correct gene order in the Lux operon-1. Pairs or primers specific for trnG/Lux-C (I), Lux-C/Lux-D (J), Lux-D/Lux-A (K), Lux-A/Lux-B (L), Lux-B/Lux-E (M), Lux-E/Lux-G (N), Lux-G/aadA (O), and aadA/trnfM (P) have been used. DNA bands (black arrows) at the predicted molecular mass (kb) confirm correct gene order. Negative controls (blanks) of PCR and DNA markers are shown in B to P.
In Vivo Validation of the MoChlo Kit
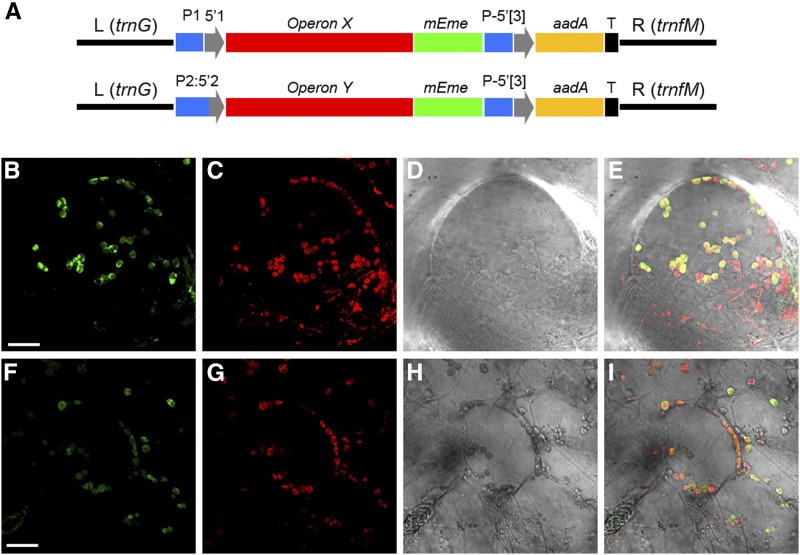
To test functionality of the MoChlo kit and demonstrate the potential of the kit for combinatorial screening, two test operons were assembled using the methods described previously. Operon X was driven by the rrn promoter, and synthetic ribosome-binding site (RBS) was used as the 5ʹUTR. For Operon Y, the fused accD promoter::5ʹUTR were used to drive expression of the operon. In both cases, the operons were assembled into Level-2 constructs for integration into the potato trnG-trnfM site (Fig. 6). In both operons, mEmerald was used as a visual marker to verify functionality of the operon by microscopy, while a monocistronic aadA cassette with the fused promoter::5ʹUTR of psbA was used for antibiotic selection of transplastomic tissue. After verification of correct assembly, the Level-2 constructs were bombarded into potato leaf discs and placed under selection with spectinomycin. After ∼10 weeks, primary green callus emerged indicating that the monocistronic aadA cassette was functional, and molecular analysis confirmed that the operons integrated correctly into the native potato plastome (Fig. 7). Further analysis of green callus by confocal microscopy showed that mEmerald fluorescence was present and correctly colocalized to the plastids in both operons (Fig. 6). In addition, the fluorescence intensity of mEmerald in Operon Y was weak compared with Operon X, which was expected due to the difference in regulatory elements between the operons. The ability to assemble, transform, and validate operons in ∼10 weeks demonstrates the potential for the complete system for combinatorial screening of plastid regulatory elements.
Figure 6.
An example of in vivo validation of two MoChlo-enabled constructs in potato chloroplasts. A, Schematic representation of test operons controlled by two examples of promoter::5ʹUTR combinations. The operon (red) is controlled by the rrn promoter (P1) along with a synthetic RBS as 5ʹUTR (5ʹ1; blue and gray; Operon X) or the promoter::5ʹUTR fusion of accD gene (P2:5ʹ2; blue and gray; Operon Y). The expression of mEmerald (mEme; green) is under the control of the operon promoter, while the selection gene, aadA (yellow), is under the control of both promoter::5ʹUTR (P-5ʹ[3]; blue and gray) and terminator/3ʹUTR (T; black) of the psbA gene. B to I, Confocal images of transplastomic green callus transformed with Operon X (B–E) or Operon Y (F–I). In both cases, the mEmerald fluorescent signal is localized to chloroplasts. B and F show mEmerald signal; C and G show chlorophyll autofluorescence; D and H show bright-field images; and E and I show merged images. Bars = 20 µm for all confocal images.
Figure 7.
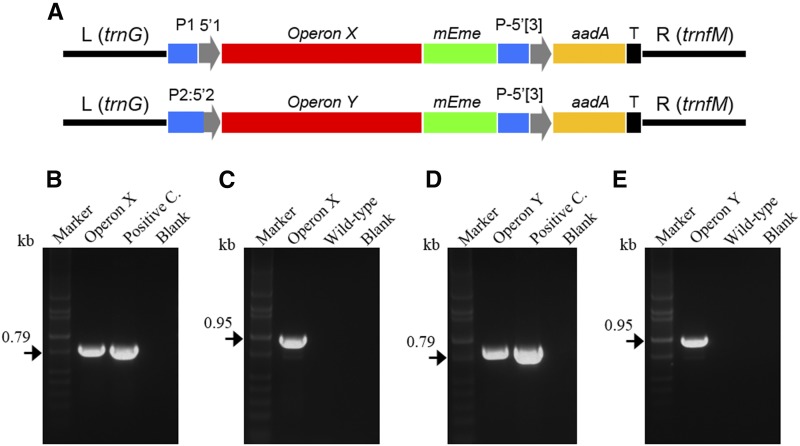
Operon integration in the potato plastome. A, Schematic representation of chloroplast constructs: rrn promoter (P1) along with a synthetic RBS used as 5′UTR (5′1; blue and gray); promoter::5′UTR fusion of accD gene (P2:5′2; blue and gray); P-5′[3], promoter::5′UTR of the psbA gene (blue and gray); T, terminator/3′UTR of psbA gene (black); operon X and Y (red); mEme (mEmerald fluorescent marker; green); and aadA (3′-adenylyltransferase selection marker; yellow). The left (L; 1.17 kb) and right (R; 1.45 kb) arms homologous to the trnG/trnfM site of the potato plastome are indicated. B to E, PCR on total genomic DNA extracted from transplastomic green callus transformed with Operon X (B and C) or Operon Y (D and E). A pair of primers specific for the aadA gene (B and D) and aadA/trnfM (C and E) have been used to check correct operon integration. DNA bands at the predicted molecular mass confirm correct integration of both operons (kb; black arrows in B–E and black line in A). Positive and negative controls (blanks) along with potato wild-type samples and DNA markers are shown.
DISCUSSION
In general, chloroplast biotechnology has fallen behind the current trends in synthetic biology and relies on antiquated systems for vector construction. To facilitate efficient assembly of large DNA constructs necessary for metabolic engineering, several molecular cloning methods based on Golden Gate assembly have been developed (Engler et al., 2008, 2009; Weber et al., 2011). Essentially, these methods take advantage of the unique ability of type IIS restriction enzymes to cut DNA outside of their recognition site, generating 4-bp-long nonpalindromic 5ʹ or 3ʹ overhangs (Engler et al., 2008, 2009). Cleavage sites can then be designed to produce compatible overhangs able to directionally assemble multiple DNA fragments in a one-pot, one-step reaction. By adding the same standardized overhangs to a library of DNA basic units, it is possible to generate a number of different combinations using the same cloning strategy (Sarrion-Perdigones et al., 2011; Weber et al., 2011; Lampropoulos et al., 2013). Compared with classical cloning techniques, Golden Gate assembly does not require a complicated design or time-consuming cloning. In fact, the combination of restriction digestion and ligation in a one-pot, one-step reaction eliminates the need for PCR amplification, fragment digestion, and gel purification, providing a fast and reliable method for assembly of large molecules of DNA (Weber et al., 2011). Golden Gate cloning can be performed in any laboratory equipped for molecular biology by simply purchasing enzymes (IIS restriction enzymes and T4 ligase), together with standardized DNA modules and vectors used for assembly (Marillonnet and Werner, 2015).
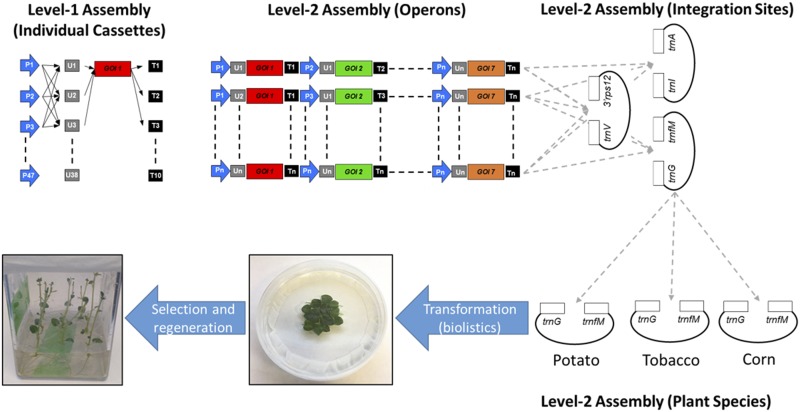
At present, only a single study has demonstrated the potential to utilize a modular cloning approach for the generation of chloroplast transformation vectors (Vafaee et al., 2014). In the previous work, GoldenBraid was chosen as the modular cloning strategy to demonstrate the potential for facile modular assembly of chloroplast transformation vectors. While GoldenBraid utilizes a smaller complement of destination/intermediate vectors, the number of parts that can be combined in a single step is more limited than with Golden Gate. As such, the MoChlo kit was designed using the Golden Gate MoClo approach. Considering the broad adoption of the plant MoClo kits, genes of interest developed by researchers for standard nuclear expression can be directly used in the MoChlo kit without further domestication. Furthermore, the MoChlo kit developed in this work has been released to the research community (through Addgene) with a complete set of Level-0 parts, including common chloroplast promoters, 5ʹUTRs, 3ʹUTRs, and selection genes (118 parts in total). Additionally, the kit contains Level-2 destination vectors specifically designed to enable homologous recombination into the most common sites for tobacco, maize, and potato chloroplast engineering. Also, the MoChlo kit has been validated in vivo, demonstrating the functionality of the components and strategy. A schematic of a combinatorial study using the MoChlo kit, from assembly of constructs to the generation of transplastomic plants, is shown in Figure 8.
Figure 8.
Schematic of the MoChlo kit for combinatorial assembly of operons for metabolic engineering. In the current iteration of the kit, combinatorial assembly of individual cassettes (Level-1 constructs) can be conducted using 47 promoters, 38 5ʹUTRs, nine promoter:5ʹUTR fusions, and 10 terminators/3ʹUTRs. In addition, the user has the ability to assemble up to seven individual cassettes, each with its own regulatory elements and GOIs. Furthermore, during Level-2 assembly, the user can choose from three different integration sites (trnI-trnA, trnV-3ʹrps12, and trnG-trnfM) from three different plant species (tobacco, maize, and potato). After completing assembly, the user would then bombard the Level-2 constructs into leaf discs and select and regenerate transplastomic plants.
Of particular importance to chloroplast biotechnology is the relative dearth of information on precise control of multigene expression. Apart from developing a fast and flexible assembly method for chloroplast transformation vectors, a secondary goal of the MoChlo kit is to accelerate research into controllable multigene expression in plastids. Therefore, promoters and regulatory sequences constitute the majority of the MoChlo kit. Optimal levels of relative protein expression between different components are fundamental to achieve successful metabolic engineering. The expression levels of protein units encoded by both monocistronic DNA assemblies and synthetic operons can be manipulated both at the transcriptional and translational levels by choosing different promoters and 5ʹUTRs, respectively. Apart from commonly used strong promoters, such as Prrn, PpsbA, and PrbcL (Shiina et al., 1998; Hayashi et al., 2003; Suzuki et al., 2003), the MoChlo kit has been supplemented with many other characterized and uncharacterized regulatory elements along with modified versions of endogenous promoters. As an example, point mutations within the −10 and −35 consensus sequences of Prrn, as well as in between them, have been shown to alter the level of transcription (Suzuki et al., 2003). Moreover, among the group of less characterized chloroplast promoters, several forms of PatpB (Xie and Allison, 2002), PatpH and PatpI (Miyagi et al., 1998), PpsbB (Tanaka et al., 1987), and PpsbD (Christopher et al., 1992; Allison and Maliga, 1995) also have been used to achieve variable gene expression. Additionally, promoters of clpP and accD have shown promise in gene expression in nongreen plastids (Zhang et al., 2012; Caroca et al., 2013). Similarly, translation motifs, such as Shine-Dalgarno sequence (or RBS) located in 5ʹUTRs, are important for efficient translation of protein. In the MoChlo kit, we provide a library of endogenous 5ʹUTRs and modified versions that are characterized by different translation efficiencies. Therefore, by using both modified versions of the same 5ʹUTR and sequences from different genes, it is possible to manipulate protein translation. For example, deletion of either psbA RBS1 or RBS2 reduces translation efficiency by ∼30%, whereas the disruption of both sequences has a drastic effect on protein translation. In contrast, deletion of the third RBS of the psbA 5ʹUTR has little effect on protein translation (Hirose and Sugiura, 1996). Similarly, mutation of the Shine-Dalgarno sequence of the rps2 5ʹUTR resulted in severalfold increase in translation efficiency compared with the wild-type sequence (Plader and Sugiura, 2003). Processed forms of 5ʹUTRs of psbN and psbB have also been shown to significantly affect translation efficiency (Yukawa et al., 2007; Kuroda and Sugiura, 2014). A chimeric synthetic 5ʹUTR generated from psbA-rbcL has also been demonstrated with a 1.5-fold decrease of translation efficiency compared with the wild-type sequences (Zou et al., 2003). Considering that the goal of many of these earlier works was to maximize transcription and translation of transgenes, many characterized regulatory elements have fallen by the wayside.
A key component for broad adoption of the MoChlo kit, and thus its usefulness to the research community, is its ease of use and efficiency. In our hands, the MoChlo kit provides a robust assembly strategy for chloroplast transformation vectors containing up to seven different cassettes. A fundamental point of emphasis for achieving high-efficiency assembly was the choice of the IIS restriction enzyme. For assembly of chloroplast transformation vectors containing only two cassettes, the choice in enzyme between BbsI and BpiI was irrelevant (Supplemental Fig. S7). For Level-2 assembly of three or more cassettes, a much higher efficiency of assembly was achieved by using BpiI rather than the isoschizomer BbsI. Attempts to assemble four- and seven-gene operons using BbsI resulted in efficiencies of 22.5% and 1.25% (Supplemental Figs. S5 and S11), compared with 92.5% and 65% using BpiI (Supplemental Figs. S6 and S10). One possible reason for the decrease in efficiency of assembly could be the kinetic properties of the two enzymes. The one-pot, one-step reaction is performed in T4 DNA ligase buffer, and we postulate that BpiI has more favorable kinetics in this suboptimal buffer. Moreover, BbsI has lower thermal stability that may reduce its activity during the Golden Gate reaction. In any event, the MoChlo kit provides a considerable advantage to small labs with limited resources and funding, in that only the genes of interest must be synthesized, and complete assembly can be accomplished using only two enzymes.
In its current form, the MoChlo kit was designed to allow assembly of seven transcriptional units into a single chloroplast transformation vector. Considering the high efficiency obtained for the maximum test case, it would be useful to identify the upper limit for efficient assembly of a chloroplast transformation vector. In a previous study, 11 Level-1 cassettes consisting of 44 Level-0 modules were assembled into a 33-kb DNA fragment using Golden Gate assembly (Weber et al., 2011). In future iterations of the MoChlo kit, the system will be expanded to assemble larger and more complex synthetic operons. For this purpose, a new set of Level-2 end linkers equipped with a visual marker (e.g. LacZα) flanked by different IIS restriction sites (BsaI or Eps3I) will be synthesized. The generation of compatible overhangs after restriction digestion will allow an additional Level-3 directional cloning to insert more transcriptional units into preassembled Level-2 constructs. Considering the relatively small size of the complete chloroplast genome of land plants (∼100–200 kb), it would theoretically be possible to synthesize a complete synthetic chloroplast genome using this approach.
CONCLUSION
The main goal of the MoChlo kit developed in this work was to provide a standardized method to assemble multigene constructs for metabolic engineering using presynthesized chloroplast destination vectors and a wide array of regulatory elements. The library of Level-0 parts developed in this work provides both commonly used and uncharacterized plastid regulatory sequences, allowing for precise modulation of multiple genes necessary to guarantee optimal protein stoichiometry for metabolic engineering. With the modularity of the approach, the MoChlo kit can be expanded easily by adding new regulatory sequences (e.g. prokaryotic inducible promoters), protein tags (e.g. fluorescent proteins and epitopes), and species-specific integration sites, along with a set of new linkers that could extend the number of transcriptional units (greater than seven) currently allowable in the kit. The substantial decrease in the cost of gene synthesis further facilitates use of the kit by researchers with limited funding and resources. The creation of a common resource of standardized Level-0 parts will provide enormous advantages in terms of collaboration and accessibility to researchers working in chloroplast biotechnology. Furthermore, the MoChlo assembly strategy is highly amenable to automation, which may further drive the potential for combinatorial screening of numerous metabolic engineering cassettes for plastid engineering.
MATERIALS AND METHODS
Design of Modules for Golden Gate Assembly
To generate Level-0 parts, the nucleotide sequences of chloroplast regulatory elements (promoters, 5ʹUTRs, and 3ʹUTRs) and genes (fluorescent/selective markers, Lux genes, and GOIs) were domesticated with both 5ʹ and 3ʹ overhangs using the criteria set forth previously (Supplemental Fig. S1; Weber et al., 2011; Engler et al., 2014). First, all IIS restriction sites (BsaI, BpiI, and Esp3I) in the native part sequences were removed by single nucleotide substitutions. Next, the parts were flanked by BsaI sites at opposite orientation along with synthetic cut sites that generate 5ʹ and 3ʹ overhangs after restriction digestion. The resulting overhangs guarantee specific directional cloning of Level-0 parts into Level-1 transcriptional units following the order promoter-5ʹUTR-GOI-3ʹUTR (Fig. 1; Supplemental Fig. S1). Compatible overhangs for Level-1 assembly and possible combinations of Level-0 parts are shown in Supplemental Figure S1. Level-0 modules of the same functional group are equipped with identical overhangs, allowing the generation of different cassettes by choosing specific modules of interest. The 5ʹ overhang of Level-0 promoters and the 3ʹ overhang of 3ʹUTRs were designed to ligate to compatible ends of Level-1 destination vectors. The overhangs used in the MoChlo kit were specifically designed to be compatible with vectors generated in previous plant MoClo systems (Engler et al., 2014). Finally, Level-0 and homologous sequences of chloroplast destination vectors were chemically synthesized by GeneArt (Invitrogen, Thermo Fisher Scientific) and cloned into Escherichia coli pUC-based vectors (kanamycin resistance gene nptII). The codon usage of Lux genes and GOI-1, GOI-2, and GOI-3 were optimized for the tobacco (Nicotiana tabacum) chloroplast genome using Optimizer (Evolutionary Genomics Group, Biochemistry and Biotechnology Department, Universitat Rovira i Virgili, Tarragona, Spain; http://genomes.urv.es/OPTIMIZER/) and a codon usage table downloaded from the codon usage database (http://www.kazusa.or.jp/codon/). Level-1 destination vectors and Level-2 end linkers from previous studies were used directly (Weber et al., 2011; Engler et al., 2014).
Golden Gate Cloning and Transformation in Escherichia coli
The one-pot, one-step reaction for Golden Gate cloning was performed in a total volume of 20 µL adding the following components in the same PCR tube. (1) Twenty-five nanograms of destination vector either for Level-1 or Level-2 and a variable amount of individual plasmids containing the fragments of interest. One hundred nanograms of plasmid containing the larger fragment along with variable amounts (ng) of other plasmids with different Mr levels. The amount of all plasmids was adjusted to guarantee the same molar ratio for all components. For Level-2, 50 ng of plasmid containing the correct end linker was also added. (2) One microliter (20 units) of the correct IIS restriction enzyme. BsaI (BsaI HF V2; New England Biolabs) was used for Level-1 and either BpiI (Anza 4 BpiI; Invitrogen, Thermo Fisher Scientific) or BbsI (BbsI HF; New England Biolabs) was used for Level-2. BpiI was used for operons larger than two cassettes. (3) One microliter (400 units) of T4 DNA ligase (New England Biolabs). (4) Two microliters of T4 DNA ligase buffer (10× stock), and the final volume was adjusted to 20 µL using sterile ultrapure water.
The Golden Gate reactions for both Level-1 and Level-2 were performed overnight in a thermocycler. A first step of 10 min at 37°C and then 10 min at 16°C was repeated for 35 cycles. The reaction was then incubated at 50°C for 30 min and 60 more min at 16°C. Before transformation into E. coli, the enzymes were inactivated for 20 min at 65°C. One microliter of Golden Gate reaction was added to 25 µL of TOP10 Chemically Competent E. coli (Invitrogen, Thermo Fisher Scientific) and transformed by the heat shock method (according to the manufacturer’s protocol). After transformation, 250 µL of super optimal broth medium (Invitrogen, Thermo Fisher Scientific) was added and then incubated for 1 h at 37°C under agitation. Fifty microliters of bacteria cultures was plated on a petri dish containing LB-agar (Luria-Bertani medium: 10 g L−1 tryptone, 10 g L−1 NaCl, 5 g L−1 yeast extract, and 15 g L−1 bacto-agar, pH 7) containing the correct selective agent. Kanamycin and spectinomycin were used at 50 and 100 µg mL−1, respectively. Liquid bacterial cultures were grown in Luria-Bertani medium without bacto-agar.
Plasmid Extraction from Bacterial Cells, Restriction Digestion, and PCR
Pure plasmid preparations were obtained using QIAprep Spin Miniprep Kit (Qiagen). The restriction digestions using either BsaI or BbsI (New England Biolabs) were performed in a total volume of 30 µL using 10 units of enzyme, 500 ng of plasmid, and 1× Custom buffer (New England Biolabs). The restriction digestion using BpiI was performed using the same conditions in Anza 1× buffer (Invitrogen, Thermo Fisher Scientific). PCRs were performed in 25-µL reaction volumes using 1× DreamTaq Green PCR Master Mix (Thermo Fisher Scientific), along with specific pairs of primers and 25 ng of template plasmid. For colony PCR, the reactions were performed using 2 µL of overnight bacteria liquid cultures. The following combinations of forward/reverse primers were used to amplify the indicated DNA fragments: GOI-1, primers 1/20; GOI-2, primers 2/21; GOI-3, primers 3/22; GOI-4, primers 4/23; aadA (full length), primers 5/24; Cassette 1A, primers 6/25; Cassette 1B and 1C, primers 7/25; Cassette 1D, primers 8/26; trnG/GOI-1, primers 9/20; GOI-1/GOI-2, primers 1/21; GOI-2/GOI-3, primers 2/22; GOI-3/aadA, primers 3/24; aadA/trnfM, primers 5/27; Lux-C, primers 10/28; Lux-D, primers 11/29; Lux-A, primers 12/30; Lux-B, primers 13/31; Lux-E, primers 14/32; Lux-G, primers 15/33; aadA (fragment), primers 16/34; mEmerald (fragment), primers 17/35; Cassette LC1, LD1, LA1, LB1, LE1, LG1, and S1, primers 18/36; trnG/Lux-C, primers 9/28; Lux-C/Lux-D, primers 10/29; Lux-D/Lux-A, primers 11/30; Lux-A/Lux-B, primers 12/31; Lux-B/Lux-E, primers 13/32; Lux-E/Lux-G, primers 14/33; Lux-G/aadA, primers 15/34; aadA/trnfM, primers 5/27; and right and left of T-DNA, primers 19/37. A list of primers used in this work is shown in Supplemental Table S4. Both restriction digestion and PCR products were separated by electrophoresis using 1% (w/v) agarose gel in 1× TAE buffer (40 mm Tris-acetate and 1 mm EDTA, pH 8.3) containing 0.3 µg mL−1 ethidium bromide. The Hi-Lo DNA ladder from 50 to 10,000 bp (BIONEXUS) was used as a molecular DNA marker.
Transformation, Selection, and Screening of Potato Chloroplasts
Potato (Solanum tuberosum var 'Desirée') was grown in sterile conditions in Magenta boxes containing MS Reg. medium (4.33 g L−1 Murashige and Skoog basal salt mixture, 25 g L−1 Suc, 100 mg L−1 myoinositol, 170 mg L−1 sodium phosphate monobasic monohydrate, 440 mg L−1 calcium chloride dihydrate, 0.9 mg L−1 thiamine-HCl, 2 mg L−1 Gly, 0.5 mg L−1 nicotinic acid, 0.5 mg L−1 pyridoxine-HCl, 1× Murashige and Skoog vitamins, and 3 g L−1 phytagel, pH 5.8). Potato plants were kept in a controlled growth room with 16 h of light (100 µmol m−2 s−1) and 8 h of dark. The temperature was kept at 24°C constant during all light/dark cycles. Transplastomic green callus was obtained from biolistic transformation of potato leaf discs as described previously (Valkov et al., 2014). Briefly, DNA constructs were bound on the surface of 600-nm gold particles (Seashell Technology) and bombarded into potato leaf discs on M6M medium. Selection of transplastomic green callus was obtained by subculturing leaf discs in M6 medium (∼4 weeks) and Ti medium (∼6 weeks) with spectinomycin, as described previously (Valkov et al., 2014).
To confirm integration, a cetyl-trimethyl-ammonium bromide-based procedure was used to extract total genomic DNA from green callus prior to confirmation of integration of the transplastomic cassette by PCR (Lin et al., 2014b). After isolation, 5 to 10 ng of total genomic DNA was used as template for PCR as described previously. The primers 5/24 and 16/27 were used to detect the full-length aadA gene and the aadA/trnfM integration region, respectively (Supplemental Table S4). Transplastomic green callus was imaged using an Olympus Fv1000 confocal microscope. The mEmerald fluorescent protein was excited at 488 nm and detected at 505 to 530 nm. Chlorophyll autofluorescence was obtained using excitation at 543 nm and detection at greater than 560 nm. Digital images were acquired using the Olympus FV10-ASW Viewer software Ver.4.2a. Confocal images were processed using ImageJ 1.41o (National Institutes of Health).
Accession Numbers
Sequences of native regulatory elements and homology arms were mined from the native plastid genomes of tobacco (Z00044.2), potato (DQ231562.1), and maize (NC_001666.2), while the E. coli terminator for rrnB (CP034595.1) and viral terminators from Tobacco mosaic virus (MK087763.1), Turnip yellow mosaic virus (KP995420.1), Brome mosaic virus (M12024.1), and Alfalfa mosaic virus (KC881009.1) were mined from their respective genomes. In addition, marker and selection genes for mEmerald (BAU94259.1), pporRFP (ABB17953.1), aadA (ARK38551.1), aphA6 (AZR92252.1), nptII (ASU89325.1), myc-aadA-GFP (AAF65230.1), FLARE16-S aadA-GFP (AAF65229.1), aac6-aph2 (WP_001028144.1), and the complete Lux operon from Photobacterium leiognathi (M63594.1) were mined from GenBank. All sequences were domesticated for use in the MoChlo kit as previously described, and the sequences of all Level-0 modules are included in Supplemental Table S1, with the entire MoChlo kit available from Addgene.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Level-0 modules and directional cloning into Level-1 cassettes.
Supplemental Figure S2. Directional cloning of Level-1 cassettes into operons of different lengths.
Supplemental Figure S3. Level-1 assembly of GOIs.
Supplemental Figure S4. Examples of Level-1 cassettes synthesized using the MoChlo kit.
Supplemental Figure S5. Colony screening for Level-2 assembly of operon-1 using BbsI in the Golden Gate reaction.
Supplemental Figure S6. Colony screening for Level-2 assembly of operon-1 using BpiI in the Golden Gate reaction.
Supplemental Figure S7. Level-2 assembly of two-gene selection cassettes using BbsI in the Golden Gate reaction.
Supplemental Figure S8. Level-2 assembly of five different four-gene operons.
Supplemental Figure S9. Level-1 assembly of Level-1 Lux cassettes.
Supplemental Figure S10. Colony screening for Level-2 assembly of Lux operon-1 using BpiI in the Golden Gate reaction.
Supplemental Figure S11. Colony screening for Level-2 assembly of Lux operon-1 using BbsI in the Golden Gate reaction.
Supplemental Table S1. MoChlo Level-0 modules.
Supplemental Table S2. Level-1 vectors compatible for the MoChlo kit.
Supplemental Table S3. Level-2 end linkers and Level-2 chloroplast destination vectors.
Supplemental Table S4. Primers used in this study.
Acknowledgments
We thank Holly Baxter and Sidney Vafaie-Partin for providing technical support throughout the project.
Footnotes
This work was supported by the Advanced Research Projects Agency-Energy (award no. DE-AR000660), USDA Hatch Grants (accession no. 1012962 and 1016740), and the Defense Advanced Research Projects Agency (award no. D17AC00016).
Articles can be viewed without a subscription.
References
- Alieva NO, Konzen KA, Field SF, Meleshkevitch EA, Hunt ME, Beltran-Ramirez V, Miller DJ, Wiedenmann J, Salih A, Matz MV (2008) Diversity and evolution of coral fluorescent proteins. PLoS ONE 3: e2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allison LA, Maliga P (1995) Light-responsive and transcription-enhancing elements regulate the plastid psbD core promoter. EMBO J 14: 3721–3730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allison LA, Simon LD, Maliga P (1996) Deletion of rpoB reveals a second distinct transcription system in plastids of higher plants. EMBO J 15: 2802–2809 [PMC free article] [PubMed] [Google Scholar]
- Apel W, Bock R (2009) Enhancement of carotenoid biosynthesis in transplastomic tomatoes by induced lycopene-to-provitamin A conversion. Plant Physiol 151: 59–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baecker JJ, Sneddon JC, Hollingsworth MJ (2009) Efficient translation in chloroplasts requires element(s) upstream of the putative ribosome binding site from atpI. Am J Bot 96: 627–636 [DOI] [PubMed] [Google Scholar]
- Bock R. (2013) Strategies for metabolic pathway engineering with multiple transgenes. Plant Mol Biol 83: 21–31 [DOI] [PubMed] [Google Scholar]
- Bock R. (2015) Engineering plastid genomes: Methods, tools, and applications in basic research and biotechnology. Annu Rev Plant Biol 66: 211–241 [DOI] [PubMed] [Google Scholar]
- Bock R, Maliga P (1995) In vivo testing of a tobacco plastid DNA segment for guide RNA function in psbL editing. Mol Gen Genet 247: 439–443 [DOI] [PubMed] [Google Scholar]
- Boyhan D, Daniell H (2011) Low-cost production of proinsulin in tobacco and lettuce chloroplasts for injectable or oral delivery of functional insulin and C-peptide. Plant Biotechnol J 9: 585–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bujarski JJ, Dreher TW, Hall TC (1985) Deletions in the 3′-terminal tRNA-like structure of brome mosaic virus RNA differentially affect aminoacylation and replication in vitro. Proc Natl Acad Sci USA 82: 5636–5640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caroca R, Howell KA, Hasse C, Ruf S, Bock R (2013) Design of chimeric expression elements that confer high-level gene activity in chromoplasts. Plant J 73: 368–379 [DOI] [PubMed] [Google Scholar]
- Carrer H, Hockenberry TN, Svab Z, Maliga P (1993) Kanamycin resistance as a selectable marker for plastid transformation in tobacco. Mol Gen Genet 241: 49–56 [DOI] [PubMed] [Google Scholar]
- Christopher DA, Kim M, Mullet JE (1992) A novel light-regulated promoter is conserved in cereal and dicot chloroplasts. Plant Cell 4: 785–798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubitt AB, Woollenweber LA, Heim R (1999) Understanding structure-function relationships in the Aequorea victoria green fluorescent protein. Methods Cell Biol 58: 19–30 [DOI] [PubMed] [Google Scholar]
- Daniell H, Datta R, Varma S, Gray S, Lee SB (1998) Containment of herbicide resistance through genetic engineering of the chloroplast genome. Nat Biotechnol 16: 345–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniell H, Chebolu S, Kumar S, Singleton M, Falconer R (2005) Chloroplast-derived vaccine antigens and other therapeutic proteins. Vaccine 23: 1779–1783 [DOI] [PubMed] [Google Scholar]
- Davoodi-Semiromi A, Schreiber M, Nalapalli S, Verma D, Singh ND, Banks RK, Chakrabarti D, Daniell H (2010) Chloroplast-derived vaccine antigens confer dual immunity against cholera and malaria by oral or injectable delivery. Plant Biotechnol J 8: 223–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eibl C, Zou Z, Beck A, Kim M, Mullet J, Koop HU (1999) In vivo analysis of plastid psbA, rbcL and rpl32 UTR elements by chloroplast transformation: Tobacco plastid gene expression is controlled by modulation of transcript levels and translation efficiency. Plant J 19: 333–345 [DOI] [PubMed] [Google Scholar]
- Emadpour M, Karcher D, Bock R (2015) Boosting riboswitch efficiency by RNA amplification. Nucleic Acids Res 43: e66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engler C, Kandzia R, Marillonnet S (2008) A one pot, one step, precision cloning method with high throughput capability. PLoS ONE 3: e3647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engler C, Gruetzner R, Kandzia R, Marillonnet S (2009) Golden gate shuffling: A one-pot DNA shuffling method based on type IIs restriction enzymes. PLoS ONE 4: e5553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engler C, Youles M, Gruetzner R, Ehnert TM, Werner S, Jones JD, Patron NJ, Marillonnet S (2014) A golden gate modular cloning toolbox for plants. ACS Synth Biol 3: 839–843 [DOI] [PubMed] [Google Scholar]
- Fuentes P, Zhou F, Erban A, Karcher D, Kopka J, Bock R (2016) A new synthetic biology approach allows transfer of an entire metabolic pathway from a medicinal plant to a biomass crop. eLife 5: e13664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammani K, Cook WB, Barkan A (2012) RNA binding and RNA remodeling activities of the half-a-tetratricopeptide (HAT) protein HCF107 underlie its effects on gene expression. Proc Natl Acad Sci USA 109: 5651–5656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi K, Shiina T, Ishii N, Iwai K, Ishizaki Y, Morikawa K, Toyoshima Y (2003) A role of the −35 element in the initiation of transcription at psbA promoter in tobacco plastids. Plant Cell Physiol 44: 334–341 [DOI] [PubMed] [Google Scholar]
- Hirata N, Yonekura D, Yanagisawa S, Iba K (2004) Possible involvement of the 5ʹ-flanking region and the 5ʹUTR of plastid accD gene in NEP-dependent transcription. Plant Cell Physiol 45: 176–186 [DOI] [PubMed] [Google Scholar]
- Hirose T, Sugiura M (1996) Cis-acting elements and trans-acting factors for accurate translation of chloroplast psbA mRNAs: Development of an in vitro translation system from tobacco chloroplasts. EMBO J 15: 1687–1695 [PMC free article] [PubMed] [Google Scholar]
- Hirose T, Sugiura M (1997) Both RNA editing and RNA cleavage are required for translation of tobacco chloroplast ndhD mRNA: A possible regulatory mechanism for the expression of a chloroplast operon consisting of functionally unrelated genes. EMBO J 16: 6804–6811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose T, Sugiura M (2004) Multiple elements required for translation of plastid atpB mRNA lacking the Shine-Dalgarno sequence. Nucleic Acids Res 32: 3503–3510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose T, Kusumegi T, Sugiura M (1998) Translation of tobacco chloroplast rps14 mRNA depends on a Shine-Dalgarno-like sequence in the 5′-untranslated region but not on internal RNA editing in the coding region. FEBS Lett 430: 257–260 [DOI] [PubMed] [Google Scholar]
- Huang FC, Klaus SM, Herz S, Zou Z, Koop HU, Golds TJ (2002) Efficient plastid transformation in tobacco using the aphA-6 gene and kanamycin selection. Mol Genet Genomics 268: 19–27 [DOI] [PubMed] [Google Scholar]
- Jin S, Daniell H (2015) The engineered chloroplast genome just got smarter. Trends Plant Sci 20: 622–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamura-Nagaya K, Ishibashi K, Huang YP, Miyashita S, Ishikawa M (2014) Replication protein of tobacco mosaic virus cotranslationally binds the 5′ untranslated region of genomic RNA to enable viral replication. Proc Natl Acad Sci USA 111: E1620–E1628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan MS, Maliga P (1999) Fluorescent antibiotic resistance marker for tracking plastid transformation in higher plants. Nat Biotechnol 17: 910–915 [DOI] [PubMed] [Google Scholar]
- Kuroda H, Maliga P (2001a) Complementarity of the 16S rRNA penultimate stem with sequences downstream of the AUG destabilizes the plastid mRNAs. Nucleic Acids Res 29: 970–975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroda H, Maliga P (2001b) Sequences downstream of the translation initiation codon are important determinants of translation efficiency in chloroplasts. Plant Physiol 125: 430–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroda H, Maliga P (2002) Overexpression of the clpP 5′-untranslated region in a chimeric context causes a mutant phenotype, suggesting competition for a clpP-specific RNA maturation factor in tobacco chloroplasts. Plant Physiol 129: 1600–1606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroda H, Sugiura M (2014) Processing of the 5′-UTR and existence of protein factors that regulate translation of tobacco chloroplast psbN mRNA. Plant Mol Biol 86: 585–593 [DOI] [PubMed] [Google Scholar]
- Kuroda H, Suzuki H, Kusumegi T, Hirose T, Yukawa Y, Sugiura M (2007) Translation of psbC mRNAs starts from the downstream GUG, not the upstream AUG, and requires the extended Shine-Dalgarno sequence in tobacco chloroplasts. Plant Cell Physiol 48: 1374–1378 [DOI] [PubMed] [Google Scholar]
- Lakshmi PS, Verma D, Yang X, Lloyd B, Daniell H (2013) Low cost tuberculosis vaccine antigens in capsules: Expression in chloroplasts, bio-encapsulation, stability and functional evaluation in vitro. PLoS ONE 8: e54708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampropoulos A, Sutikovic Z, Wenzl C, Maegele I, Lohmann JU, Forner J (2013) GreenGate: A novel, versatile, and efficient cloning system for plant transgenesis. PLoS ONE 8: e83043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CY, Szittner RB, Meighen EA (1991) The lux genes of the luminous bacterial symbiont, Photobacterium leiognathi, of the ponyfish: Nucleotide sequence, difference in gene organization, and high expression in mutant Escherichia coli. Eur J Biochem 201: 161–167 [DOI] [PubMed] [Google Scholar]
- Liere K, Maliga P (1999) In vitro characterization of the tobacco rpoB promoter reveals a core sequence motif conserved between phage-type plastid and plant mitochondrial promoters. EMBO J 18: 249–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin MT, Occhialini A, Andralojc PJ, Devonshire J, Hines KM, Parry MA, Hanson MR (2014a) β-Carboxysomal proteins assemble into highly organized structures in Nicotiana chloroplasts. Plant J 79: 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin MT, Occhialini A, Andralojc PJ, Parry MA, Hanson MR (2014b) A faster Rubisco with potential to increase photosynthesis in crops. Nature 513: 547–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long BM, Hee WY, Sharwood RE, Rae BD, Kaines S, Lim YL, Nguyen ND, Massey B, Bala S, von Caemmerer S, et al. (2018) Carboxysome encapsulation of the CO2-fixing enzyme Rubisco in tobacco chloroplasts. Nat Commun 9: 3570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Rijzaani H, Karcher D, Ruf S, Bock R (2013) Efficient metabolic pathway engineering in transgenic tobacco and tomato plastids with synthetic multigene operons. Proc Natl Acad Sci USA 110: E623–E632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz KA, Azhagiri AK, Tungsuchat-Huang T, Maliga P (2007) A guide to choosing vectors for transformation of the plastid genome of higher plants. Plant Physiol 145: 1201–1210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J, Pallett D, Jiang H, Hou Y, Wang H (2015) Mutational bias of Turnip Yellow Mosaic Virus in the context of host anti-viral gene silencing. Virology 486: 2–6 [DOI] [PubMed] [Google Scholar]
- Maliga P. (2004) Plastid transformation in higher plants. Annu Rev Plant Biol 55: 289–313 [DOI] [PubMed] [Google Scholar]
- Maliga P, Bock R (2011) Plastid biotechnology: Food, fuel, and medicine for the 21st century. Plant Physiol 155: 1501–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marillonnet S, Werner S (2015) Assembly of multigene constructs using Golden Gate cloning. Methods Mol Biol 1321: 269–284 [DOI] [PubMed] [Google Scholar]
- McNeal JR, Kuehl JV, Boore JL, de Pamphilis CW (2007) Complete plastid genome sequences suggest strong selection for retention of photosynthetic genes in the parasitic plant genus Cuscuta. BMC Plant Biol 7: 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyagi T, Kapoor S, Sugita M, Sugiura M (1998) Transcript analysis of the tobacco plastid operon rps2/atpI/H/F/A reveals the existence of a non-consensus type II (NCII) promoter upstream of the atpI coding sequence. Mol Gen Genet 257: 299–307 [DOI] [PubMed] [Google Scholar]
- Occhialini A, Lin MT, Andralojc PJ, Hanson MR, Parry MA (2016) Transgenic tobacco plants with improved cyanobacterial Rubisco expression but no extra assembly factors grow at near wild-type rates if provided with elevated CO2. Plant J 85: 148–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oey M, Lohse M, Kreikemeyer B, Bock R (2009a) Exhaustion of the chloroplast protein synthesis capacity by massive expression of a highly stable protein antibiotic. Plant J 57: 436–445 [DOI] [PubMed] [Google Scholar]
- Oey M, Lohse M, Scharff LB, Kreikemeyer B, Bock R (2009b) Plastid production of protein antibiotics against pneumonia via a new strategy for high-level expression of antimicrobial proteins. Proc Natl Acad Sci USA 106: 6579–6584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasoreck EK, Su J, Silverman IM, Gosai SJ, Gregory BD, Yuan JS, Daniell H (2016) Terpene metabolic engineering via nuclear or chloroplast genomes profoundly and globally impacts off-target pathways through metabolite signalling. Plant Biotechnol J 14: 1862–1875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plader W, Sugiura M (2003) The Shine-Dalgarno-like sequence is a negative regulatory element for translation of tobacco chloroplast rps2 mRNA: An additional mechanism for translational control in chloroplasts. Plant J 34: 377–382 [DOI] [PubMed] [Google Scholar]
- Sarrion-Perdigones A, Falconi EE, Zandalinas SI, Juárez P, Fernández-del-Carmen A, Granell A, Orzaez D (2011) GoldenBraid: An iterative cloning system for standardized assembly of reusable genetic modules. PLoS ONE 6: e21622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schelkunov MI, Shtratnikova VY, Nuraliev MS, Selosse MA, Penin AA, Logacheva MD (2015) Exploring the limits for reduction of plastid genomes: A case study of the mycoheterotrophic orchids Epipogium aphyllum and Epipogium roseum. Genome Biol Evol 7: 1179–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindel HS, Piatek AA, Stewart CN Jr, Lenaghan SC (2018) The plastid genome as a chassis for synthetic biology-enabled metabolic engineering: Players in gene expression. Plant Cell Rep 37: 1419–1429 [DOI] [PubMed] [Google Scholar]
- Shaver JM, Oldenburg DJ, Bendich AJ (2006) Changes in chloroplast DNA during development in tobacco, Medicago truncatula, pea, and maize. Planta 224: 72–82 [DOI] [PubMed] [Google Scholar]
- Shiina T, Allison L, Maliga P (1998) rbcL transcript levels in tobacco plastids are independent of light: Reduced dark transcription rate is compensated by increased mRNA stability. Plant Cell 10: 1713–1722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sriraman P, Silhavy D, Maliga P (1998) The phage-type PclpP-53 plastid promoter comprises sequences downstream of the transcription initiation site. Nucleic Acids Res 26: 4874–4879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staub JM, Maliga P (1992) Long regions of homologous DNA are incorporated into the tobacco plastid genome by transformation. Plant Cell 4: 39–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staub JM, Maliga P (1994) Translation of psbA mRNA is regulated by light via the 5′-untranslated region in tobacco plastids. Plant J 6: 547–553 [DOI] [PubMed] [Google Scholar]
- Suzuki JY, Sriraman P, Svab Z, Maliga P (2003) Unique architecture of the plastid ribosomal RNA operon promoter recognized by the multisubunit RNA polymerase in tobacco and other higher plants. Plant Cell 15: 195–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svab Z, Maliga P (1993) High-frequency plastid transformation in tobacco by selection for a chimeric aadA gene. Proc Natl Acad Sci USA 90: 913–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabatabaei I, Ruf S, Bock R (2017) A bifunctional aminoglycoside acetyltransferase/phosphotransferase conferring tobramycin resistance provides an efficient selectable marker for plastid transformation. Plant Mol Biol 93: 269–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M, Obokata J, Chunwongse J, Shinozaki K, Sugiura M (1987) Rapid splicing and stepwise processing of a transcript from the psbB operon in tobacco chloroplasts: Determination of the intron sites in petB and petD. Mol Gen Genet 209: 427–431 [DOI] [PubMed] [Google Scholar]
- Tangphatsornruang S, Birch-Machin I, Newell CA, Gray JC (2011) The effect of different 3′ untranslated regions on the accumulation and stability of transcripts of a gfp transgene in chloroplasts of transplastomic tobacco. Plant Mol Biol 76: 385–396 [DOI] [PubMed] [Google Scholar]
- Tregoning JS, Nixon P, Kuroda H, Svab Z, Clare S, Bowe F, Fairweather N, Ytterberg J, van Wijk KJ, Dougan G, et al. (2003) Expression of tetanus toxin Fragment C in tobacco chloroplasts. Nucleic Acids Res 31: 1174–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trucco V, de Breuil S, Bejerman N, Lenardon S, Giolitti F (2014) Complete nucleotide sequence of Alfalfa mosaic virus isolated from alfalfa (Medicago sativa L.) in Argentina. Virus Genes 48: 562–565 [DOI] [PubMed] [Google Scholar]
- Vafaee Y, Staniek A, Mancheno-Solano M, Warzecha H (2014) A modular cloning toolbox for the generation of chloroplast transformation vectors. PLoS ONE 9: e110222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valkov VT, Gargano D, Scotti N, Cardi T (2014) Plastid transformation in potato: Solanum tuberosum. Methods Mol Biol 1132: 295–303 [DOI] [PubMed] [Google Scholar]
- Verhounig A, Karcher D, Bock R (2010) Inducible gene expression from the plastid genome by a synthetic riboswitch. Proc Natl Acad Sci USA 107: 6204–6209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma D, Daniell H (2007) Chloroplast vector systems for biotechnology applications. Plant Physiol 145: 1129–1143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma D, Samson NP, Koya V, Daniell H (2008) A protocol for expression of foreign genes in chloroplasts. Nat Protoc 3: 739–758 [DOI] [PubMed] [Google Scholar]
- Weber E, Engler C, Gruetzner R, Werner S, Marillonnet S (2011) A modular cloning system for standardized assembly of multigene constructs. PLoS ONE 6: e16765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurbs D, Ruf S, Bock R (2007) Contained metabolic engineering in tomatoes by expression of carotenoid biosynthesis genes from the plastid genome. Plant J 49: 276–288 [DOI] [PubMed] [Google Scholar]
- Xie G, Allison LA (2002) Sequences upstream of the YRTA core region are essential for transcription of the tobacco atpB NEP promoter in chloroplasts in vivo. Curr Genet 41: 176–182 [DOI] [PubMed] [Google Scholar]
- Yukawa M, Kuroda H, Sugiura M (2007) A new in vitro translation system for non-radioactive assay from tobacco chloroplasts: Effect of pre-mRNA processing on translation in vitro. Plant J 49: 367–376 [DOI] [PubMed] [Google Scholar]
- Zhang J, Ruf S, Hasse C, Childs L, Scharff LB, Bock R (2012) Identification of cis-elements conferring high levels of gene expression in non-green plastids. Plant J 72: 115–128 [DOI] [PubMed] [Google Scholar]
- Zhou F, Karcher D, Bock R (2007) Identification of a plastid intercistronic expression element (IEE) facilitating the expression of stable translatable monocistronic mRNAs from operons. Plant J 52: 961–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou F, Badillo-Corona JA, Karcher D, Gonzalez-Rabade N, Piepenburg K, Borchers AM, Maloney AP, Kavanagh TA, Gray JC, Bock R (2008) High-level expression of human immunodeficiency virus antigens from the tobacco and tomato plastid genomes. Plant Biotechnol J 6: 897–913 [DOI] [PubMed] [Google Scholar]
- Zou Z, Eibl C, Koop HU (2003) The stem-loop region of the tobacco psbA 5'UTR is an important determinant of mRNA stability and translation efficiency. Mol Genet Genomics 269: 340–349 [DOI] [PubMed] [Google Scholar]