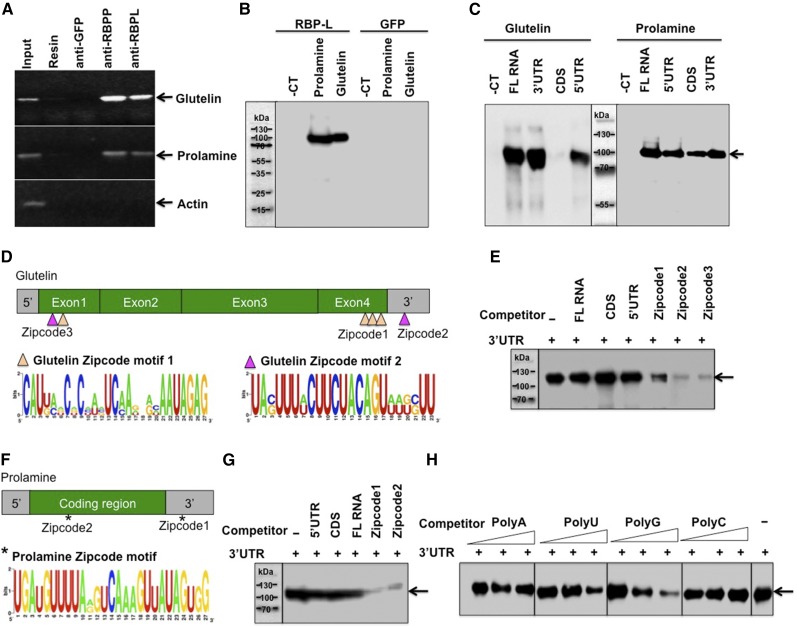
Figure 2.
RBP-L binds to glutelin and prolamine mRNAs, especially to their zipcode RNA sequences. A, RNA-IP analysis showing that RBP-L is associated with glutelin and prolamine mRNAs in vivo. The RNAs extracted from IPs generated by anti-RBP-P, anti-RBP-L, anti-GFP, and empty resin were subjected to RT-PCR to detect glutelin, prolamine, and actin gene sequences. The figure depicts an agarose gel with resolved PCR products. Anti-GFP and empty resin were used as negative controls. Input, PCR using cDNA synthesized from RNAs isolated from IPs. B, Binding activity of RBP-L to glutelin and prolamine mRNAs compared to RNA control (for detailed information, see “Materials and Methods”). The figure depicts an immunoblot where anti-DIG antibody was used to detect bound DIG-labeled RNAs (see “Materials and Methods” for detailed experimental details). GFP protein was used as negative control for the binding test. C, Binding activities of RBP-L to different region of glutelin and prolamine RNAs. Arrow indicates the location of recombinant RBP-L. D, The glutelin RNA zipcode elements, which consist of two types of motifs, zipcode motif 1 (orange triangle) and zipcode motif 2 (magenta triangle). (Top) Location and components of three glutelin zipcode elements; (bottom) consensus zipcode sequences generated by WebLogo (http://weblogo.berkeley.edu). E, Competition assays using equimolar amounts of DIG-labeled glutelin 3′UTR and the indicated unlabeled competitor RNAs. Arrows indicates the location of recombinant RBP-L. F, The prolamine zipcode elements, which consist of only a single zipcode motif (*). (Top) Location of the two zipcode elements within prolamine mRNA; (bottom) consensus sequence of prolamine zipcode. G, Competition assays using equimolar amounts of DIG-labeled prolamine 3′UTR and indicated unlabeled competitor RNAs. Arrows indicate the location of recombinant RBP-L. H, Binding specificity of RBP-L to different homoribopolymers. Unlabeled poly(U), poly(A), poly(C), and poly(G) were added at 5-, 10-, and 20-fold mole excess over DIG-labeled glutelin 3′ UTR RNA. Arrows indicates the location of recombinant RBP-L. -CT, pBlueScript KS vector sequence; 5′, 5′UTR; 3′, 3′UTR; -, no competitor.