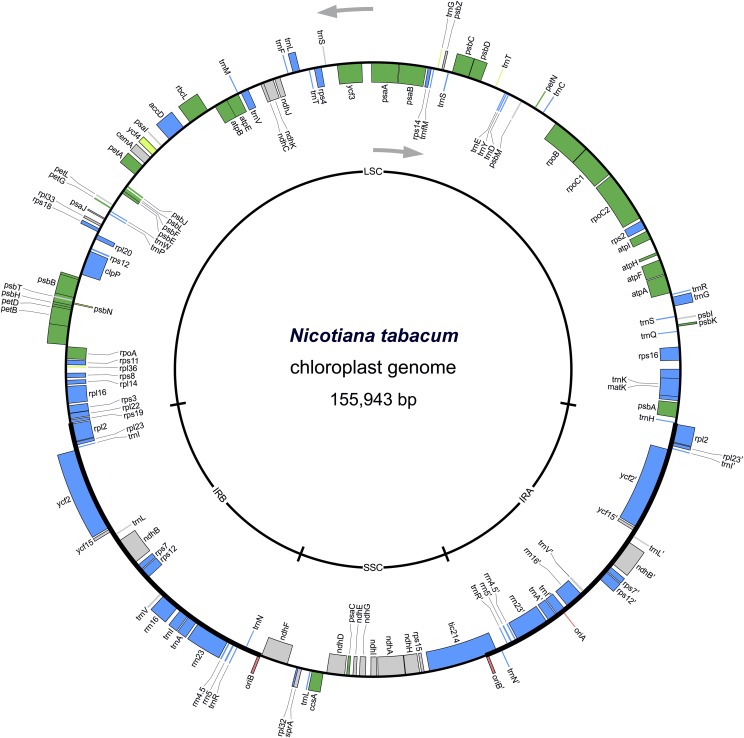
Figure 3.
Physical map of the N. tabacum chloroplast genome with all genes classified by essentiality. Genes shown in blue are essential for both heterotrophic and autotrophic growth. Genes shown in green are essential for autotrophic growth only. Light green indicates borderline cases where knock-out plants survive under carefully controlled growth conditions. Genes shown in gray are nonessential under both heterotrophic and autotrophic growth conditions, in that their knock-out causes no or only a mild phenotype (Scharff and Bock, 2014). Origins of replication are highlighted in red. Gray arrows indicate the direction of transcription for the two DNA strands. The map was drawn using the OrganellarGenomeDRAW (OGDRAW) software (Lohse et al., 2013) based on the complete plastome sequence of N. tabacum (Shinozaki et al., 1986; GenBank accession number Z00044.2). LSC, large single-copy region; IRA, inverted repeat A; IRB, inverted repeat B; SSC, small single-copy region.