Abstract
The environmental Alphaproteobacterium Caulobacter crescentus is a classical model to study the regulation of the bacterial cell cycle. It divides asymmetrically, giving a stalked cell that immediately enters S phase and a swarmer cell that stays in the G1 phase until it differentiates into a stalked cell. Its genome consists in a single circular chromosome whose replication is tightly regulated so that it happens only in stalked cells and only once per cell cycle. Imbalances in chromosomal copy numbers are the most often highly deleterious, if not lethal. This review highlights recent discoveries on pathways that control chromosome replication when Caulobacter is exposed to optimal or less optimal growth conditions. Most of these pathways target two proteins that bind directly onto the chromosomal origin: the highly conserved DnaA initiator of DNA replication and the CtrA response regulator that is found in most Alphaproteobacteria. The concerted inactivation and proteolysis of CtrA during the swarmer-to-stalked cell transition license cells to enter S phase, while a replisome-associated Regulated Inactivation and proteolysis of DnaA (RIDA) process ensures that initiation starts only once per cell cycle. When Caulobacter is stressed, it turns on control systems that delay the G1-to-S phase transition or the elongation of DNA replication, most probably increasing its fitness and adaptation capacities.
Keywords: chromosomes, Caulobacter, CtrA, DNA replication and recombination, DnaA, Gram-negative bacteria
Introduction
DNA replication is controlled with exquisite precision in all cell types to ensure that each daughter cell inherits one copy of complete chromosome(s) following each division event. Control mechanisms largely focus on the initiation step of the replication process, when the replisome is assembled onto DNA. In nearly all bacteria, the highly conserved initiator of chromosome replication is DnaA [1,2]. It typically binds to several DnaA boxes located on the chromosomal origin and oligomerizes to open the DNA double helix at an AT-rich region within the origin. It also interacts with helicases to load them onto the DNA. Subsequently, the replicative DNA polymerase and its β-sliding clamp (DnaN) are recruited onto leading and lagging strands to initiate bi-directional and processive DNA replication. The proper timing of chromosome replication is highly dependent on controlling the levels, the activity and the availability of DnaA in bacteria. The analysis of a variety of bacteria now reveals to which extent control mechanisms are conserved, or not, in different bacterial classes or species [3,4]. It also shows that bacteria often control the DNA replication process to modulate their proliferation in response to environmental cues directly connected to their biological niche, through the regulation of DnaA or of other more specific regulators.
Caulobacter crescentus (henceforth Caulobacter) is an aquatic Alphaproteobacterium that emerged as a powerful model system to study the regulation of the bacterial cell cycle. This bacterium divides asymmetrically at the end of each cell cycle giving daughter cells with distinct developmental and replicative fates (Figure 1) [5]. The first one is a replication incompetent (G1-phase) swarmer cell, while the second one is a replication competent (S-phase) stalked cell. To initiate the replication of its unique circular chromosome, the swarmer cell must first differentiate into a stalked cell. This relatively slow growing bacterium, compared with the most studied Escherichia coli model system, never displays more than two replication forks at work within the same cell and over-initiation events are severely deleterious [6]. It is relatively easy to isolate nearly pure populations of swarmer cells from mixed populations of Caulobacter cells, facilitating studies on the regulation of the timing of DNA replication during the bacterial cell cycle.
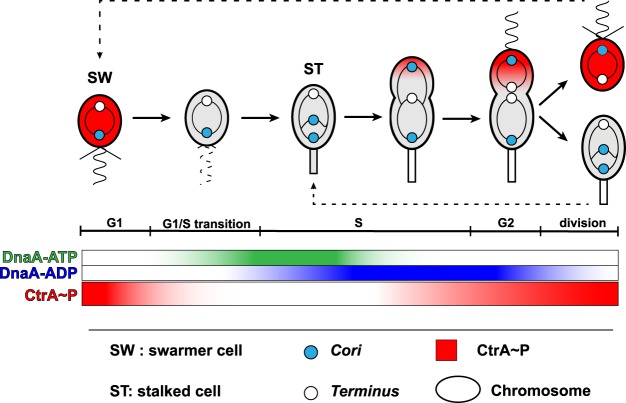
Figure 1. Graphical representation of the Caulobacter cell cycle and of the abundancy of the DnaA and CtrA main regulators of the initiation of chromosomal replication.
In swarmer cells, the initiation of chromosomal replication is inhibited by high levels of CtrA∼P (red color) binding to the chromosomal origin of replication (Cori, blue circle). During the swarmer-to-stalked cell transition, CtrA∼P is proteolyzed at the cell pole where the stalk will be built. Coincidentally, DnaA is synthesized and binds to ATP. DnaA-ATP (green color) binds to the Cori and is active to initiate replication. As soon as DNA replication has started, HdaA is recruited to the replisome and activated to stimulate to conversion of DnaA-ATP into DnaA-ADP (blue color) and DnaA proteolysis. This Regulated Inactivation of DnaA (RIDA) process prevents re-initiation during the same cell cycle. In early predivisional cells, CtrA is re-synthesized and phosphorylated, but gets efficiently dephosphorylated and proteolyzed in the swarmer cell compartment of late pre-divisional cells to inhibit the initiation of DNA replication in the swarmer progeny. Although this was not directly demonstrated, CtrA∼P may form a gradient in pre-divisional cells, being the most abundant at the flagellated cell pole even before cell compartmentalization, due to polarized upstream regulators of CtrA phosphorylation and degradation [83].
This review focuses on recently discovered mechanisms that control the replication of the chromosome of Caulobacter cultivated in favorable or less-favorable growth conditions, with particular emphasis on the regulation of the DnaA initiator and of the CtrA inhibitor of chromosome replication that are both highly conserved in Alphaproteobacteria.
The origin of replication of the Caulobacter crescentus chromosome
The origin of replication of the Caulobacter chromosome (Cori) was mapped years ago at a chromosomal locus located close to the hemE and overlapping the CCNA_00001 (duf299) open reading frames (ORF) (Figure 2) [6–8]. It carries DNA motifs with affinity for several known regulators of the Caulobacter cell cycle, including the initiator DnaA, the response regulator CtrA, the nucleoid associated proteins IHF (Integration host Factor) and GapR, and the DNA methyltransferase (MTase) CcrM [6,7,9]. As assumed in nearly all bacterial species, DnaA, and several of the DnaA boxes found on the origin, are indispensable for the initiation of chromosome replication in Caulobacter [7,10]. DnaA binds to seven DnaA boxes on the Cori; five of these are low-affinity W-boxes and two are moderate-affinity G-boxes (Figure 2) [7]. Although one of the W-box overlaps with a DNA motif that can be methylated by the DNA MTase CcrM (Figure 2), it appears that methylation of the Cori by CcrM is not required for DNA replication in Caulobacter, since the chromosome content of ΔccrM cells is identical with that of wild-type cells [11,12]. Also, consistent with this finding, a homolog of the methylation-dependent DNA binding protein SeqA, which controls replication initiation in E. coli [3], cannot be found in the Caulobacter proteome. The relatively low affinity of DnaA for the Cori DnaA boxes might be connected with the existence of CtrA, which inhibits the initiation, supposedly through a direct competition with DnaA when binding to the Cori. Indeed, five CtrA binding sites are found on the Cori and one of them overlaps one of the two moderate-affinity DnaA G-boxes (Figure 2). The ctrA gene is essential for the survival of Caulobacter and partial loss of CtrA function leads to premature initiation of DNA replication in swarmer cells [13,14]. Notably, an IHF binding site overlaps one of the CtrA binding sites (Figure 2) [15], indicating that IHF may promote the disassembly of CtrA on the Cori during the swarmer-to-stalked cell transition [16]. Targeted mutagenesis experiments eliminating the CtrA binding sites on the Cori, however, showed that the inhibition of DNA replication by CtrA is dispensible when Caulobacter is cultivated in minimal medium [17]. In addition to DnaA, CtrA and IHF, the GapR protein shows affinity for Cori [9]. GapR is a newly discovered and conserved nucleoid associated protein that appears as critical for chromosome replication and growth in Caulobacter [9,18,19], most probably by promoting the ability of topoisomerases to relax positive supercoils that accumulate ahead of the replication fork [20]. Considering that GapR is more abundant in stalked than in swarmer cells [9], it might also promote the binding of DnaA to the Cori, although this was not tested directly. Finally, the presence of two ORFs [21] and of a gene transcribed into a small non-coding RNA [22,23] in the Cori region (Figure 2) suggests that the transcription of these elements could potentially interfere with or promote the opening of the DNA double helix for initiation. Consistent with this possibility, early findings showed that transcription from one of the hemE promoters is required for chromosome replication in Caulobacter [24].
Figure 2. Organization of the Caulobacter chromosomal origin of replication (Cori).
The Cori carries two classes of DnaA binding sites: G-boxes (G1 and G2; orange arrows) of moderate affinity and W-boxes (W1–W5; green arrows) of low affinity. Note that the G2 box overlaps one of the CtrA binding sites (red squares), suggesting that DnaA-ATP may compete with CtrA when binding to the Cori. The Cori also carries an IHF binding site (blue square) and five GANTC sites (fuchsia color; –CH3) that are methylated by the CcrM DNA methyltransferase. IHF may promote the opening of the DNA double helix at the AT-rich region (purple box) when DnaA-ATP is bound to the Cori. In addition, the transcriptional start sites (TSS; represented by a thin black arrow) for three RNAs are located in the Cori: the duf299 (CCNA_00001) and hemE mRNAs and the CCNA_R0094 small non-coding RNA. Transcription starting at these TSS might influence initiation.
CtrA restricts the initiation of chromosome replication to stalked cells
CtrA is a conserved response regulator, which is found in most Alphaproteobacteria [25]. It needs to be phosphorylated to be active and to inhibit the initiation of DNA replication [13]. Logically, very strict regulatory mechanisms control the levels of active CtrA∼P (Figures 1 and 3A), to ensure that chromosome replication can still start once per cell cycle and specifically during the swarmer-to-stalked cell transition of Caulobacter or in the stalked progeny following cell division. CtrA∼P is very abundant in swarmer cells, where it binds to the Cori with high affinity to maintain cells in G1 phase, and in pre-divisional cells, where it plays essential roles in the regulation of gene expression (Figure 1) [26,27]. Following cytoplasmic compartmentalization in late pre-divisional cell, the levels of CtrA become highly dissimilar: the flagellated compartment of the cell contains ∼22,000 molecules of CtrA [28], while the stalked compartment contains undetectable levels of CtrA. These strong temporal and spatial variations are mostly dependent on the activity of the essential CckA histidine kinase/phosphatase, which is at the top of a phosphorelay controlling CtrA phosphorylation and proteolysis (Figure 3A) [5,6]. Indeed, although ctrA transcription varies significantly during the cell cycle, due to tight control by the GcrA epigenetic regulator [29,30], by the CtrA-associated SciP regulator [31,32] and by self-regulation [33], this temporal regulation is not required for the control of the initiation of replication since the constitutive transcription of ctrA does not lead to replication defects [26]. Still, sufficient transcription of ctrA is needed for the correct control of replication initiation, since a loss of CtrA activity leads to severe replication defects and cell death [13].
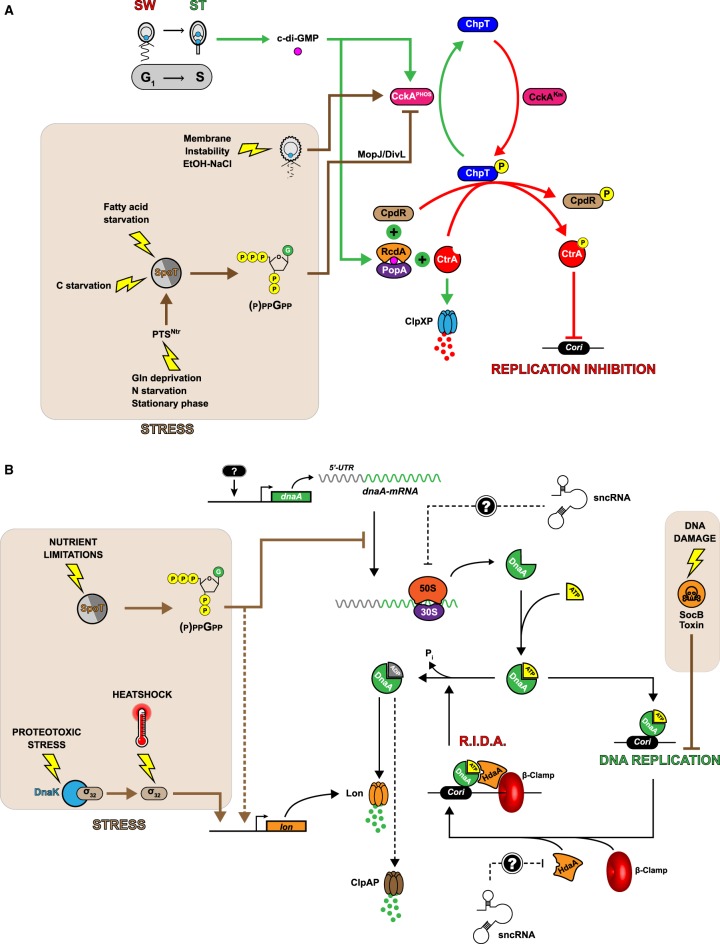
Figure 3. Graphical representation of the regulatory circuits controlling CtrA and DnaA activity in Caulobacter cells cultivated in optimal (no background color) or stressful (brown background color) growth conditions.
(A) Model for the regulatory network controlling the levels and the activity of CtrA. During the swarmer-to-stalked cell transition, the activity of CckA switches from a kinase (CckAKIN; red arrows) to a phosphatase (CckAPHOS; green arrows), reducing the levels of ChpT∼P. In turn, CtrA∼P can no more accumulate and gets actively degraded by a CpdR- and PopA/RcdA-dependent ClpXP proteolysis complex, triggering the initiation of DNA replication by DnaA-ATP. When nutrient levels become too limiting, the kinase activity of CckA is probably stimulated in a (p)ppGpp- and MopJ-dependent manner to delay the G1-to-S phase transition. In contrast, membrane stresses appear to promote the phosphatase activity of CckA and a reduction in CtrA∼P levels, maybe to delay cell division without inhibiting chromosome replication. (B) Model of the regulatory network controlling the levels and the activity of DnaA. Newly synthesized DnaA associates with ATP, which can then initiate DNA replication once CtrA∼P levels are low enough. Immediately after replication has started active DnaA-ATP is converted into inactive DnaA-ADP by the β-clamp binding HdaA protein. This conversion also appears to promote the degradation of DnaA by the Lon protease. This Regulated Inactivation and proteolysis of DnaA (RIDA) process ensures that DNA replication cannot re-initiate before the next cell cycle. Another β-clamp binding protein named SocB can inhibit the elongation of DNA replication in response to DNA damage. When Caulobacter is exposed to stresses such as nutrient limitations or proteotoxic stresses, DnaA levels drop due to an inhibition of dnaA translation and/or to a stimulation of DnaA proteolysis.
CckA is the most important regulator of CtrA levels and activity. It acts as a kinase phosphorylating the ChpT phosphotransferase in swarmer and pre-divisional cells, while it acts as a phosphatase in stalked cells (Figure 3A). In turn, phosphorylated ChpT transfers its phosphate group to the two response regulators CtrA and CpdR, activating and stabilizing CtrA [34,35]. The switch in CckA activity from a kinase to a phosphatase mode is controlled by the cyclic diguanylate (c-di-GMP) second messenger (Figure 3A) [36,37], whose levels rise specifically during the swarmer-to-stalked cell transition and in the swarmer compartment of late pre-divisional cells [38] and which interacts directly with one of the two PAS domains of CckA [39]. In addition, this PAS domain interacts with the DivL pseudokinase in complex with the DivK response regulator to further control the activity of CckA [40–42]. The kinase-to-phosphatase switch in CckA activity dictates when the initiation of chromosome replication can start, so that it takes place specifically during the swarmer-to-stalked cell transition or in the newborn stalked cell following division [14]. In addition to inactivating CtrA by de-phosphorylation, this switch also promotes CtrA degradation by the ClpXP protease by de-phosphorylating CpdR (Figure 3A) [34,35]. When CpdR is un-phosphorylated, it associates with ClpX and with a complex composed of the c-di-GMP-stimulated PopA protein and of the RcdA adaptor to turn on CtrA proteolysis at the cell pole [43–47]. Although CtrA degradation takes place at the stalked pole of wild-type cells, recent findings, however, suggest that it can take place even more efficiently in the cytoplasm of cells lacking the PopZ cell pole organizer [48,49]. Once the replication of the chromosome is licensed to start through the strong reduction in CtrA∼P levels, DnaA can bind to the Cori to initiate DNA replication.
DnaA controls the frequency of the initiation of chromosome replication
Control of the abundancy of active DnaA ensures that replication can start once, but only once, per cell cycle [6,50]. Levels must be high enough in stalked cells, but shut down right after replication initiation to prevent over-initiation events in stalked and early pre-divisional cells, when CtrA∼P levels are minimal.
A first level of regulation is through the control of dnaA transcription that peaks right before the initiation of chromosome replication during the swarmer-to-stalked cell transition [6,51]. Mechanisms controlling dnaA transcription are still unclear although the analysis of dnaA promoter elements indicates that the efficiency and the timing of dnaA transcription may be controlled by a transcriptional activator binding close to a GAGTC motif upstream of the −35 region [52,53]. A second level of regulation is through the inhibition of dnaA translation by a long untranslated region (5'UTR) upstream of the translational start codon (Figure 3B), but this mechanism does not appear to control the timing of dnaA expression [53]. Importantly, the transcriptional and post-transcriptional controls of DnaA synthesis play only a marginal role in controlling when, and at which frequency, DNA replication can initiate during the Caulobacter cell cycle, since the artificial and constitutive expression of dnaA does not lead to replication defects in Caulobacter. Instead, these mechanisms may be more useful to control DnaA levels in response to environmental signals, rather than to cell cycle cues.
The most important mechanism controlling the frequency of replication initiation is the so-called Regulated Inactivation of DnaA (RIDA) process that appears as essential for the survival of Caulobacter [14,54,55]. This process restricts the levels of active DnaA by stimulating its ATPase activity, leading to the concerted inactivation and degradation of DnaA (Figure 3B) [6,50]. There are now clear indications that DnaA needs to be associated with ATP to initiate DNA replication [14,54,55], as it is also the case in many other bacterial species [3]. The RIDA process takes place right after the initiation of DNA replication, through a stimulation of the ATPase activity of DnaA by the HdaA protein, preventing lethal over-initiation events. The current model is that HdaA interacts with the β-sliding clamp of the replisome once it is loaded onto the DNA at the onset of DNA replication and that this interaction switches on the activity of HdaA and the subsequent RIDA process [54,56]. In addition, the conversion of DnaA-ATP into DnaA-ADP during the RIDA process leads to a significant destabilization of DnaA at the onset of DNA replication during the swarmer-to-stalked cell transition [57,58]. Two different ATP-dependent proteases can recognize DnaA in Caulobacter (Figure 3B). The first one is ClpAP, which appears to destabilize DnaA independently of the nucleotide bound to it and preferentially under stress conditions, since the ClpS adaptor inhibits this pathway in exponentially growing cells [59]. The second one is Lon [60] and seems to have a preference for DnaA-ADP rather than DnaA-ATP [59], suggesting that it preferentially degrades DnaA during the S phase of the cell cycle when the RIDA process is active. Thus, Lon probably contributes to controlling the intracellular levels of active DnaA as a function of the cell cycle to prevent over-initiation events. As described below, it also plays a key role in adjusting DnaA levels and regulating DNA replication in response to stresses.
Control of chromosome replication during non-optimal growth conditions
Bacteria must coordinate the replication of their genome with their growth rate. This is all the more important for bacteria that live in oligotrophic environments, like Caulobacter, and that are thus frequently exposed to nutrient limitations and environmental stresses.
When exposed to nutrient limitations, Caulobacter turns on a so-called stringent response that targets DnaA and CtrA to slow down or arrest the replication of its chromosome. This response is based on the production of guanosine tetra- or penta-phosphate ((p)ppGpp) by the SpoT enzyme [61], in response to carbon or nitrogen starvation [62] or in response to fatty acid depletion (Figure 3A) [63]. Recent findings also showed that SpoT is specifically activated by elements of the nitrogen-related phosphotransferase system (PTSNtr) in response to low glutamine levels [64,65]. The (p)ppGpp alarmone then appears to inhibit DnaA synthesis (Figure 3B) and CtrA degradation (Figure 3A), leading to a severe inhibition of the initiation of chromosome replication [66]. The stabilization of CtrA by (p)ppGpp appears to be dependent on the up-regulation of the conserved MopJ regulator, which targets the DivL-dependent pathway controlling CckA activity (Figure 3A), although this may not be the only pathway involved [67]. The complex regulatory system controlling DnaA levels in response to nutrient limitations appears to include an inhibition of dnaA translation (Figure 3B), leading to a rapid clearance of DnaA by the Lon protease [68]. Interestingly, this translational inhibition also takes place in starved ΔspoT mutant cells, indicating that other unknown (p)ppGpp-independent pathways are also involved in the regulation of DNA replication in response to low nutrient levels [68].
When exposed to DNA/protein damaging conditions, Caulobacter also stops or slows down the replication of its chromosome. Two such regulatory pathways have now been identified (Figure 3B). The first one is based on the detection of unfolded or damaged proteins by the DnaK chaperone, which leads to a stabilization of the σ32 heat-shock factor and the consequent activation of lon transcription and Lon-mediated degradation of DnaA [60]. Thus, proteotoxic and severe heat-shock stresses block the initiation of DNA replication by DnaA [60,69]. The second one is based on the stabilization of the SocB toxin in response to DNA damage. In turn, SocB can bind to the β-clamp of the DNA polymerase to inhibit the elongation of DNA replication, most probably through a disassembly of the replicative DNA polymerase complex [70].
Surprisingly, Caulobacter adopts a rather opposite strategy when facing stress conditions affecting the integrity of its membrane, such as exposure to ethanol or high salt concentrations in its environment or exposure to mild heat-shocks. Indeed, these stresses promote the phosphatase activity of CckA in a DivL- and c-di-GMP-independent manner, leading a rapid decrease in the levels of active CtrA∼P [71]. As a consequence, fewer stressed cells are in G1 phase and cells appear longer with an abnormally high DNA content compared with non-stressed cells. Future work should aim at understanding why the inhibition of CtrA activity provides an advantage to cells facing membrane stresses, while it appears to be deleterious to cells facing nutritional stresses.
Perspectives
This review aimed at summarizing the current view on the control of DNA replication in Caulobacter. Most, although not all, of the pathways regulating chromosome replication target the DnaA initiator or the CtrA response regulator. In fast growing conditions, the tight regulation of their activity is mostly dependent on the RIDA process and on the activity of the CckA kinase/phosphatase. A common point between the RIDA process and the CckA phosphatase is that they promote the inactivation and the proteolysis of their targets (Figure 3). Interestingly, the (p)ppGpp alarmone affects CtrA and DnaA to adjust the length of the G1 phase under fast-growing conditions [72], or to block cells in the G1 phase under stressful conditions [64,65,67]. DnaA synthesis also appears to be regulated by (p)ppGpp and growth conditions [66,68], but mechanisms involved in these control systems are still ill-defined. Interestingly, experimental and computational evidences suggest that up to 10 small non-coding RNAs (sncRNAs) might target the dnaA and hdaA messenger RNAs (Figure 3B) [22,23,73]. Considering that the expression of several of these is cell cycle-regulated or regulated in response to stresses [22,23,74,75], and that the Hfq RNA chaperone is required for metabolic homeostasis [76], it is tempting to predict that translational regulation by sncRNAs might also play an important role in fine-tuning the levels and the activity of DnaA (Figure 3B). Other unknown pathways are probably involved in adjusting DNA replication with cell growth, most probably by connecting the central metabolism with DNA replication. A recent study, for example, raised the possibility that a member of the Lrp family of transcription factors, named PutR, might connect proline intracellular levels with the elongation of DNA replication and cell division in Caulobacter [77].
It is noteworthy that DnaA and CtrA are also important transcription factors [27,78]. The impact of the RIDA process on the control of DnaA-regulated genes has not been investigated in detail, but preliminary findings suggest that the nucleotide binding to DnaA might also influence the transcription of a subset of these genes and chromosome segregation [55,79]. This might be used by Caulobacter to coordinate DNA replication with other processes required for cell cycle progression.
Finally, it is also interesting to mention that while the regulation of DnaA appears to be a widespread mechanism used by most bacteria to control chromosome replication, the involvement of CtrA in replication control is restricted to only a subset of them. Indeed, although CtrA is well conserved in Alphaproteobacteria, its regulon appears to evolve rapidly [80–82]. Then, determining whether DNA replication is under negative control in other Alphaproteobacteria and understanding the associated mechanisms could provide interesting new information on the evolution of complex networks controlling chromosome replication in bacteria.
Acknowledgements
We thank Giulia Cheloni for the critical reading of this manuscript. We apologize to authors not cited in this short mini-review, which mostly focuses on articles published over the last five years.
Funding
The Swiss National Science Foundation provided financial support (SNSF fellowships CRSII3_160703 and 31003A_173075).
Competing Interests
The Authors declare that there are no competing interests associated with the manuscript.
References
- 1.Katayama T., Ozaki S., Keyamura K. and Fujimitsu K. (2010) Regulation of the replication cycle: conserved and diverse regulatory systems for DnaA and oriC. Nat. Rev. Microbiol. 8, 163–170 10.1038/nrmicro2314 [DOI] [PubMed] [Google Scholar]
- 2.Katayama T., Kasho K. and Kawakami H. (2017) The DnaA cycle in Escherichia coli: activation, function and inactivation of the initiator protein. Front. Microbiol. 8, 2496 10.3389/fmicb.2017.02496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Skarstad K. and Katayama T. (2013) Regulating DNA replication in bacteria. Cold Spring Harb. Perspect. Biol. 5, a012922 10.1101/cshperspect.a012922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Heinrich K., Leslie D.J. and Jonas K. (2015) Modulation of bacterial proliferation as a survival strategy. Adv. Appl. Microbiol. 92, 127–171 10.1016/bs.aambs.2015.02.004 [DOI] [PubMed] [Google Scholar]
- 5.Collier J. (2016) Cell cycle control in Alphaproteobacteria. Curr. Opin. Microbiol. 30, 107–113 10.1016/j.mib.2016.01.010 [DOI] [PubMed] [Google Scholar]
- 6.Collier J. (2012) Regulation of chromosomal replication in Caulobacter crescentus. Plasmid 67, 76–87 10.1016/j.plasmid.2011.12.007 [DOI] [PubMed] [Google Scholar]
- 7.Taylor J.A., Ouimet M.C., Wargachuk R. and Marczynski G.T. et al. (2011) The Caulobacter crescentus chromosome replication origin evolved two classes of weak DnaA binding sites. Mol. Microbiol. 82, 312–326 10.1111/j.1365-2958.2011.07785.x [DOI] [PubMed] [Google Scholar]
- 8.Marczynski G.T. and Shapiro L. (1992) Cell-cycle control of a cloned chromosomal origin of replication from Caulobacter crescentus. J. Mol. Biol. 226, 959–977 10.1016/0022-2836(92)91045-Q [DOI] [PubMed] [Google Scholar]
- 9.Taylor J.A., Panis G., Viollier P.H. and Marczynski G.T. (2017) A novel nucleoid-associated protein coordinates chromosome replication and chromosome partition. Nucleic Acids Res. 45, 8916–8929 10.1093/nar/gkx596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gorbatyuk B. and Marczynski G.T. (2001) Physiological consequences of blocked Caulobacter crescentus dnaA expression, an essential DNA replication gene. Mol. Microbiol. 40, 485–497 10.1046/j.1365-2958.2001.02404.x [DOI] [PubMed] [Google Scholar]
- 11.Gonzalez D., Kozdon J.B., McAdams H.H., Shapiro L. and Collier J. (2014) The functions of DNA methylation by CcrM in Caulobacter crescentus: a global approach. Nucleic Acids Res. 42, 3720–3735 10.1093/nar/gkt1352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mouammine A. and Collier J. (2018) The impact of DNA methylation in Alphaproteobacteria. Mol. Microbiol. 110, 1–10 10.1111/mmi.14079 [DOI] [PubMed] [Google Scholar]
- 13.Quon K.C., Yang B., Domian I.J., Shapiro L. and Marczynski G.T. (1998) Negative control of bacterial DNA replication by a cell cycle regulatory protein that binds at the chromosome origin. Proc. Natl Acad. Sci. U.S.A. 95, 120–125 10.1073/pnas.95.1.120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jonas K., Chen Y.E. and Laub M.T. (2011) Modularity of the bacterial cell cycle enables independent spatial and temporal control of DNA replication. Curr. Biol. 21, 1092–1101 10.1016/j.cub.2011.05.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Siam R., Brassinga A.K. and Marczynski G.T. (2003) A dual binding site for integration host factor and the response regulator CtrA inside the Caulobacter crescentus replication origin. J. Bacteriol. 185, 5563–5572 10.1128/JB.185.18.5563-5572.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Muir R.E. and Gober J.W. (2005) Role of integration host factor in the transcriptional activation of flagellar gene expression in Caulobacter crescentus. J. Bacteriol. 187, 949–960 10.1128/JB.187.3.949-960.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bastedo D.P. and Marczynski G.T. (2009) Ctra response regulator binding to the Caulobacter chromosome replication origin is required during nutrient and antibiotic stress as well as during cell cycle progression. Mol. Microbiol. 72, 139–154 10.1111/j.1365-2958.2009.06630.x [DOI] [PubMed] [Google Scholar]
- 18.Arias-Cartin R., Dobihal G.S., Campos M., Surovtsev I.V, Parry B. and Jacobs-Wagner C. (2017) Replication fork passage drives asymmetric dynamics of a critical nucleoid-associated protein in Caulobacter. EMBO J. 36, 301–318 10.15252/embj.201695513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ricci D.P., Melfi M.D., Lasker K., Dill D.L., McAdams H.H. and Shapiro L. (2016) Cell cycle progression in Caulobacter requires a nucleoid-associated protein with high AT sequence recognition. Proc. Natl. Acad. Sci. U.S.A. 113, E5952–E5961 10.1073/pnas.1612579113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guo M.S., Haakonsen D.L., Zeng W., Schumacher M.A. and Laub M.T. (2018) A bacterial chromosome structuring protein binds overtwisted DNA to stimulate type II topoisomerases and enable DNA replication. Cell 175, 583–597.e23 10.1016/j.cell.2018.08.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shaheen S.M., Ouimet M.-C. and Marczynski G.T. (2009) Comparative analysis of Caulobacter chromosome replication origins. Microbiology 155, 1215–1225 10.1099/mic.0.025528-0 [DOI] [PubMed] [Google Scholar]
- 22.Schrader J.M., Zhou B., Li G.-W., Lasker K., Childers W.S., Williams B. et al. (2014) The coding and noncoding architecture of the Caulobacter crescentus genome. PLoS Genet. 10, e1004463 10.1371/journal.pgen.1004463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou B., Schrader J.M., Kalogeraki V.S., Abeliuk E., Dinh C.B., Pham J.Q. et al. (2015) The global regulatory architecture of transcription during the Caulobacter cell cycle. PLoS Genet. 11, e1004831 10.1371/journal.pgen.1004831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marczynski G.T., Lentine K. and Shapiro L. (1995) A developmentally regulated chromosomal origin of replication uses essential transcription elements. Genes Dev. 9, 1543–1557 10.1101/gad.9.12.1543 [DOI] [PubMed] [Google Scholar]
- 25.Brilli M., Fondi M., Fani R., Mengoni A., Ferri L., Bazzicalupo M. et al. (2010) The diversity and evolution of cell cycle regulation in alpha-proteobacteria: a comparative genomic analysis. BMC Syst. Biol. 4, 52 10.1186/1752-0509-4-52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Domian I.J., Quon K.C. and Shapiro L. (1997) Cell type-specific phosphorylation and proteolysis of a transcriptional regulator controls the G1-to-S transition in a bacterial cell cycle. Cell 90, 415–424 10.1016/S0092-8674(00)80502-4 [DOI] [PubMed] [Google Scholar]
- 27.Laub M.T., Chen S.L., Shapiro L. and McAdams H.H. (2002) Genes directly controlled by CtrA, a master regulator of the Caulobacter cell cycle. Proc. Natl Acad. Sci. U.S.A. 99, 4632–4637 10.1073/pnas.062065699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Judd E.M., Ryan K.R., Moerner W.E., Shapiro L. and McAdams H.H. (2003) Fluorescence bleaching reveals asymmetric compartment formation prior to cell division in Caulobacter. Proc. Natl Acad. Sci. U.S.A. 100, 8235–8240 10.1073/pnas.1433105100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Holtzendorff J., Hung D., Brende P., Reisenauer A., Viollier1 P.H., McAdams H.H. et al. (2004) Oscillating global regulators control the genetic circuit driving a bacterial cell cycle. Science 304, 983–987 10.1126/science.1095191 [DOI] [PubMed] [Google Scholar]
- 30.Fioravanti A., Fumeaux C., Mohapatra S.S., Bompard C., Brilli M., Frandi A. et al. (2013) DNA binding of the cell cycle transcriptional regulator GcrA depends on N6-Adenosine methylation in Caulobacter crescentus and other alphaproteobacteria. PLoS Genet. 9, e1003541 10.1371/journal.pgen.1003541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tan M.H., Kozdon J.B., Shen X., Shapiro L. and McAdams H.H. (2010) An essential transcription factor, SciP, enhances robustness of Caulobacter cell cycle regulation. Proc. Natl Acad. Sci. U.S.A. 107, 18985–18990 10.1073/pnas.1014395107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gora K.G., Tsokos C.G., Chen Y.E., Srinivasan B.S., Perchuk B.S. and Laub M.T. (2010) A cell-type-specific protein-protein interaction modulates transcriptional activity of a master regulator in Caulobacter crescentus. Mol. Cell 39, 455–467 10.1016/j.molcel.2010.06.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Domian I.J., Reisenauer A. and Shapiro L. (1999) Feedback control of a master bacterial cell-cycle regulator. Proc. Natl Acad. Sci. U.S.A. 96, 6648–6653 10.1073/pnas.96.12.6648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Biondi E.G. Reisinger S.J., Skerker J.M., Arif M., Perchuk B.S., Ryan K.R. et al. (2006) Regulation of the bacterial cell cycle by an integrated genetic circuit. Nature 444, 899–904 10.1038/nature05321 [DOI] [PubMed] [Google Scholar]
- 35.Iniesta A.A., McGrath P.T., Reisenauer A., McAdams H.H. and Shapiro L. (2006) A phospho-signaling pathway controls the localization and activity of a protease complex critical for bacterial cell cycle progression. Proc. Natl Acad. Sci. U.S.A. 103, 10935–10940 10.1073/pnas.0604554103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lori C., Ozaki S., Steiner S., Böhm R., Abel S., Dubey B.N. et al. (2015) Cyclic di-GMP acts as a cell cycle oscillator to drive chromosome replication. Nature 523, 236–239 10.1038/nature14473 [DOI] [PubMed] [Google Scholar]
- 37.Dubey B.N., Lori C., Ozaki S., Fucile G., Plaza-Menacho I., Jenal U. et al. (2016) Cyclic di-GMP mediates a histidine kinase/phosphatase switch by noncovalent domain cross-linking. Sci. Adv. 2, e1600823 10.1126/sciadv.1600823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Abel S., Bucher T., Nicollier M., Hug I., Kaever V., Abel zur Wiesch P. et al. (2013) Bi-modal distribution of the second messenger c-di-GMP controls cell fate and asymmetry during the Caulobacter cell cycle. PLoS Genet. 9, e1003744 10.1371/journal.pgen.1003744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mann T.H., Seth Childers W., Blair J.A., Eckart M.R. and Shapiro L. (2016) A cell cycle kinase with tandem sensory PAS domains integrates cell fate cues. Nat. Commun. 7, 11454 10.1038/ncomms11454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Childers W.S., Xu Q., Mann T.H., Mathews I.I., Blair J.A., Deacon A.M. et al. (2014) Cell fate regulation governed by a repurposed bacterial histidine kinase. PLoS Biol. 12, e1001979 10.1371/journal.pbio.1001979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Childers W.S. and Shapiro L. (2014) A pseudokinase couples signaling pathways to enable asymmetric cell division in a bacterium. Microb. Cell 2, 29–32 10.15698/mic2015.01.184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mann T.H. and Shapiro L. (2018) Integration of cell cycle signals by multi-PAS domain kinases. Proc. Natl Acad. Sci. U.S.A. 115, E7166–E7173 10.1073/pnas.1808543115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smith S.C., Joshi K.K., Zik J.J., Trinh K., Kamajaya A., Chien P. et al. (2014) Cell cycle-dependent adaptor complex for ClpXP-mediated proteolysis directly integrates phosphorylation and second messenger signals. Proc. Natl Acad. Sci. U.S.A. 111, 14229–14234 10.1073/pnas.1407862111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ozaki S., Schalch-Moser A., Zumthor L., Manfredi P., Ebbensgaard A., Schirmer T. et al. (2014) Activation and polar sequestration of PopA, a c-di-GMP effector protein involved in Caulobacter crescentus cell cycle control. Mol. Microbiol. 94, 580–594 10.1111/mmi.12777 [DOI] [PubMed] [Google Scholar]
- 45.Lau J., Hernandez-Alicea L., Vass R.H. and Chien P. (2015) A phosphosignaling adaptor primes the AAA+ protease ClpXP to drive cell cycle-regulated proteolysis. Mol. Cell 59, 104–116 10.1016/j.molcel.2015.05.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Joshi K.K., Bergé M., Radhakrishnan S.K., Viollier P.H. and Chien P. (2015) An adaptor hierarchy regulates proteolysis during a bacterial cell cycle. Cell 163, 419–431 10.1016/j.cell.2015.09.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Joshi K.K., Sutherland M. and Chien P. (2017) Cargo engagement protects protease adaptors from degradation in a substrate-specific manner. J. Biol. Chem. 292, 10973–10982 10.1074/jbc.M117.786392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Joshi K.K., Battle C.M. and Chien P. (2018) Polar localization Hub protein PopZ restrains adaptor-Dependent ClpXP proteolysis in Caulobacter crescentus. J. Bacteriol. 200 10.1128/JB.00221-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bergé M. and Viollier P.H. (2018) End-in-sight: cell polarization by the polygamic organizer PopZ. Trends. Microbiol. 26, 363–375 10.1016/j.tim.2017.11.007 [DOI] [PubMed] [Google Scholar]
- 50.Felletti M., Omnus D.J. and Jonas K. (2018) Regulation of the replication initiator dnaA in Caulobacter crescentus. Biochim. Biophys. Acta Gene. Regul. Mech. 10.1016/j.bbagrm.2018.01.004 [DOI] [PubMed] [Google Scholar]
- 51.Zweiger G. and Shapiro L. (1994) Expression of Caulobacter dnaA as a function of the cell cycle. J. Bacteriol. 176, 401–408 10.1128/jb.176.2.401-408.1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Collier J., McAdams H.H. and Shapiro L. (2007) A DNA methylation ratchet governs progression through a bacterial cell cycle. Proc. Natl Acad. Sci. U.S.A. 104, 17111–6 10.1073/pnas.0708112104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cheng L. and Keiler K.C. (2009) Correct timing of dnaA transcription and initiation of DNA replication requires trans translation. J. Bacteriol. 191, 4268–4275 10.1128/JB.00362-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Collier J. and Shapiro L. (2009) Feedback control of DnaA-mediated replication initiation by replisome-associated HdaA protein in Caulobacter. J. Bacteriol. 191, 5706–5716 10.1128/JB.00525-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fernandez-Fernandez C., Gonzalez D. and Collier J. (2011) Regulation of the activity of the dual-function DnaA protein in Caulobacter crescentus. PLoS ONE 6, e26028 10.1371/journal.pone.0026028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fernandez-Fernandez C., Grosse K., Sourjik V. and Collier J. (2013) The beta-sliding clamp directs the localization of HdaA to the replisome in Caulobacter crescentus. Microbiology 159, 2237–2248 10.1099/mic.0.068577-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wargachuk R. and Marczynski G.T. (2015) The Caulobacter crescentus homolog of DnaA (HdaA) also regulates the proteolysis of the replication initiator protein DnaA. J. Bacteriol. 197, 3521–3532 10.1128/JB.00460-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gorbatyuk B. and Marczynski G.T. (2005) Regulated degradation of chromosome replication proteins DnaA and CtrA in Caulobacter crescentus. Mol. Microbiol. 55, 1233–1245 10.1111/j.1365-2958.2004.04459.x [DOI] [PubMed] [Google Scholar]
- 59.Liu J., Francis L.I., Jonas K., Laub M.T. and Chien P. (2016) ClpAP is an auxiliary protease for DnaA degradation in Caulobacter crescentus. Mol. Microbiol. 102, 1075–1085 10.1111/mmi.13537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jonas K., Liu J., Chien P. and Laub M.T. (2013) Proteotoxic stress induces a cell-cycle arrest by stimulating Lon to degrade the replication initiator DnaA. Cell 154, 623–636 10.1016/j.cell.2013.06.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lesley J.A. and Shapiro L. (2008) Spot regulates DnaA stability and initiation of DNA replication in carbon-starved Caulobacter crescentus. J. Bacteriol. 190, 6867–6880 10.1128/JB.00700-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Boutte C.C. and Crosson S. (2011) The complex logic of stringent response regulation in Caulobacter crescentus: starvation signalling in an oligotrophic environment. Mol. Microbiol. 80, 695–714 10.1111/j.1365-2958.2011.07602.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stott K.V., Wood S.M., Blair J.A., Nguyen B.T., Herrera A., Mora Y.G.P. et al. (2015) (P)ppGpp modulates cell size and the initiation of DNA replication in Caulobacter crescentus in response to a block in lipid biosynthesis. Microbiology 161(Pt), 553–564 10.1099/mic.0.000032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ronneau S., Petit K., De Bolle X. and Hallez R. (2016) Phosphotransferase-dependent accumulation of (p)ppGpp in response to glutamine deprivation in Caulobacter crescentus. Nat. Commun. 7, 11423 10.1038/ncomms11423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sanselicio S. and Viollier P.H. (2015) Convergence of alarmone and cell cycle signaling from Trans-encoded sensory domains. MBio 6 10.1128/mBio.01415-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gonzalez D. and Collier J. (2014) Effects of (p)ppGpp on the progression of the cell cycle of Caulobacter crescentus. J. Bacteriol. 196, 2514–2525 10.1128/JB.01575-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sanselicio S., Bergé M., Théraulaz L., Radhakrishnan S.K. and Viollier P.H. (2015) Topological control of the Caulobacter cell cycle circuitry by a polarized single-domain PAS protein. Nat. Commun. 6, 7005 10.1038/ncomms8005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Leslie D.J., Heinen C., Schramm F.D., Thüring M., Aakre C.D., Murray S.M. et al. (2015) Nutritional control of DNA replication initiation through the proteolysis and regulated translation of dnaA. PLoS Genet. 11, e1005342 10.1371/journal.pgen.1005342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schramm F.D., Heinrich K., Thüring M., Bernhardt J., Jonas K. and Viollier P.H. (2017) An essential regulatory function of the DnaK chaperone dictates the decision between proliferation and maintenance in Caulobacter crescentus. PLoS Genet. 13, e1007148 10.1371/journal.pgen.1007148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Aakre C.D., Phung T.N., Huang D. and Laub M.T. (2013) A bacterial toxin inhibits DNA replication elongation through a direct interaction with the beta sliding clamp. Mol. Cell 52, 617–628 10.1016/j.molcel.2013.10.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Heinrich K., Sobetzko P. and Jonas K. (2016) A kinase-phosphatase switch transduces environmental information into a bacterial cell cycle circuit. PLoS Genet. 12, e1006522 10.1371/journal.pgen.1006522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Boutte C.C., Henry J.T. and Crosson S. (2012) Ppgpp and polyphosphate modulate cell cycle progression in Caulobacter crescentus. J. Bacteriol. 194, 28–35 10.1128/JB.05932-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Beroual W., Brilli M. and Biondi E.G. (2018) Non-coding RNAs potentially controlling cell cycle in the model Caulobacter crescentus: a bioinformatic approach. Front. Genet. 9, 164 10.3389/fgene.2018.00164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Landt S.G., Abeliuk E., McGrath P.T., Lesley J.A., McAdams H.H. and Shapiro L. (2008) Small non-coding RNAs in Caulobacter crescentus. Mol. Microbiol. 68, 600–614 10.1111/j.1365-2958.2008.06172.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lasker K., Schrader J.M., Men Y., Marshik T., Dill D.L., McAdams H.H. et al. (2016) Caulobrowser: a systems biology resource for Caulobacter crescentus. Nucleic Acids Res. 44, D640–D645 10.1093/nar/gkv1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Irnov I., Wang Z., Jannetty N.D., Bustamante J.A., Rhee K.Y., Jacobs-Wagner C. et al. (2017) Crosstalk between the tricarboxylic acid cycle and peptidoglycan synthesis in Caulobacter crescentus through the homeostatic control of alpha-ketoglutarate. PLoS Genet. 13, e1006978 10.1371/journal.pgen.1006978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mouammine A., Eich K., Frandi A. and Collier J. (2018) Control of proline utilization by the Lrp-like regulator PutR in Caulobacter crescentus. Sci. Rep. 8, 14677 10.1038/s41598-018-32660-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hottes A.K., Shapiro L. and McAdams H.H. (2005) Dnaa coordinates replication initiation and cell cycle transcription in Caulobacter crescentus. Mol. Microbiol. 58, 1340–1353 10.1111/j.1365-2958.2005.04912.x [DOI] [PubMed] [Google Scholar]
- 79.Mera P.E., Kalogeraki V.S. and Shapiro L. (2014) Replication initiator dnaA binds at the Caulobacter centromere and enables chromosome segregation. Proc. Natl Acad. Sci. U.S.A. 111, 16100–5 10.1073/pnas.1418989111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Pini F., De Nisco N.J., Ferri L., Penterman J., Fioravanti A., Brilli M. et al. (2015) Cell cycle control by the master regulator CtrA in Sinorhizobium meliloti. PLoS Genet. 11, e1005232 10.1371/journal.pgen.1005232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Figueroa-Cuilan W., Daniel J.J., Howell M., Sulaiman A. and Brown P.J.B. (2016) Mini-Tn7 insertion in an artificial attTn7 site enables depletion of the essential master regulator ctrA in the phytopathogen agrobacterium tumefaciens. Appl. Environ. Microbiol. 82, 5015–5025 10.1128/AEM.01392-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Francis N., Poncin K., Fioravanti A., Vassen V., Willemart K., Ong T.A.P. et al. (2017) Ctra controls cell division and outer membrane composition of the pathogen Brucella abortus. Mol. Microbiol. 103, 780–797 10.1111/mmi.13589 [DOI] [PubMed] [Google Scholar]
- 83.Tsokos C.G. and Laub M.T. (2012) Polarity and cell fate asymmetry in Caulobacter crescentus. Curr. Opin. Microbiol. 15, 744–750 10.1016/j.mib.2012.10.011 [DOI] [PMC free article] [PubMed] [Google Scholar]