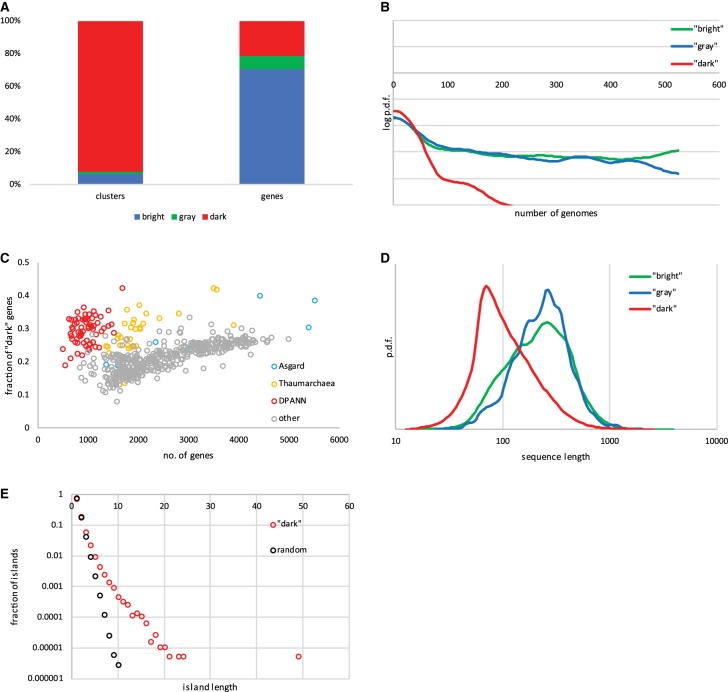
Figure 1. Dark matter in archaeal genomes.
Amino acid sequences of proteins, encoded in 524 (nearly) completely sequenced archaeal genomes were, when possible, assigned to 13 443 arCOGs [16] and the rest were clustered together. The combination of arCOGs and clusters is referred to as ‘gene families' here and elsewhere in the text. (A) The relative frequencies of ‘dark' (no functional annotation), ‘gray' (general functional prediction only), and ‘bright' (functionally annotated) matter among archaeal gene families (arCOGs and clusters) and individual genes. (B) Distribution of the number of genomes represented in the ‘dark', ‘gray', and ‘bright matter’ gene families. The plot shows the Gaussian kernel smoothed probability density functions in log scale; the number of genomes ranges from 1 (ORFan gene) to 524 (strictly ubiquitous family). (C) The fraction of ‘dark matter' genes in 524 archaeal genomes. (D) Distribution of the sequence lengths among the ‘dark’, ‘gray', and ‘bright matter' protein families. The plot shows the Gaussian kernel smoothed probability density functions, calculated for the family consensus sequences. (E) Distribution of the island lengths (lengths of contiguous blocks of genes) for the ‘dark matter' genes and for a randomly selected gene set of the same size (285 155 genes).