Abstract
Mediator is a large multiprotein complex conserved in all eukaryotes that plays an essential role in transcriptional regulation. Mediator comprises 25 subunits in yeast and 30 subunits in humans that form three main modules and a separable four-subunit kinase module. For nearly 20 years, because of its size and complexity, Mediator has posed a formidable challenge to structural biologists. The first two-dimensional electron microscopy (EM) projection map of Mediator leading to the canonical view of its division in three topological modules named Head, Middle and Tail, was published in 1999. Within the last few years, optimization of Mediator purification combined with technical and methodological advances in cryo-electron microscopy (cryo-EM) have revealed unprecedented details of Mediator subunit organization, interactions with RNA polymerase II and parts of its core structure at high resolution. To celebrate the twentieth anniversary of the first Mediator EM reconstruction, we look back on the structural studies of Mediator complex from a historical perspective and discuss them in the light of our current understanding of its role in transcriptional regulation.
Keywords: Cryo-EM, Mediator complex, structural biology, transcription
Introduction
The Mediator complex was initially identified in yeast in the early 1990s independently by the Kornberg and the Young laboratories. The Kornberg laboratory isolated Mediator biochemically from a distinct crude yeast fraction that stimulated activator-dependent transcription in vitro using partially purified proteins [1,2]. The Young laboratory used yeast genetic screens and identified the first Mediator genes as suppressors of truncations of the Carboxy-terminal domain (CTD) of RNA polymerase II (RNA Pol II). The product of four dominants suppressors termed Srb2, Srb4, Srb5 and Srb6 (Srb, suppressor of RNA polymerase B [3]) were shown to be part of a high molecular mass multisubunit complex that was tightly bound to the RNA Pol II [4]. An activity was soon after isolated that stimulated transcription in vitro in a form of a 20-subunit complex including Srb2, Srb4, Srb5 and Srb6 [5] (reviewed in [6]). Complexes with similar activities were subsequently purified in metazoans by many laboratories (reviewed in [7]). At first, it was unclear whether these complexes were all related to the yeast Mediator but comparative genomics [8,9] and multidimensional protein identification technology (MudPIT) [10] identified a set of consensus Mediator subunits conserved in all eukaryotes and a unified nomenclature was adopted in 2004 [11].
Mediator acts as a physical and functional bridge between DNA-binding transcription factors and the transcription machinery. It regulates gene expression at multiple stages of transcription, from promoting assembly of the preinitiation complex (PIC) to facilitating efficient entry into elongation or promoter escape (reviewed in [12–19]). Owing to its large size, its multisubunit composition, its conformational flexibility and the presence of numerous intrinsically disordered regions in many subunits [20,21], it remains very challenging to determine the complete structure of Mediator at high resolution. Initial EM investigations of negatively stained Mediator preparations provided outlines of the overall architecture of the complex as well as of the Mediator–Pol II holoenzyme complex at low resolution. These studies identified different modules within Mediator, referred to as Head, Middle and Tail in yeast [22–24] and initially named Head, body and leg in humans [25]. Mediator complexes can be isolated at least as two distinct stable entities containing or lacking the 4-subunit cyclin-dependent kinase 8 (CDK8) kinase module [25]. Over the past two decades, particularly in the last couple of years, remarkable progresses have been made in understanding structure–function relationships for Mediator as well as its role in the PIC, especially in yeast (reviewed in [26–30]). Here, we review recent insights regarding the Mediator complex structure and place them in historical perspective (Figure 1 and Supplementary Table S1).
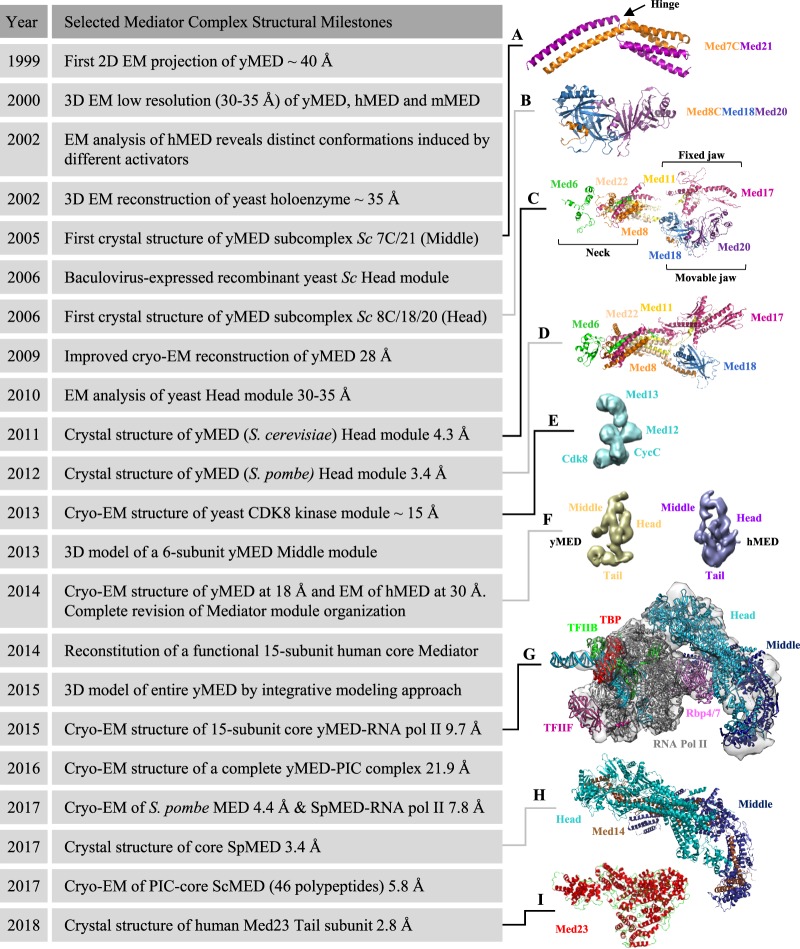
Figure 1. Timeline of selected milestones in Mediator complex structural studies.
From the top down, the depicted structures are (A) S. cerevisiae Med7C/Med21 complex (protein data bank (PDB) 1YKH [42]). The conserved flexible hinge is indicated. (B) S. cerevisiae Med8C/18/20 submodule (PDB 2HZS [43]), (C) S. cerevisiae Head module (PDB 3RJ1 [53]). The three major domains described initially in 2011 (fixed jaw, movable jaw and neck) are indicated. (D) S. pombe Head module (PDB 4H63 [55]), (E) S. cerevisiae CDK8 kinase module (electron microscopy data bank (EMD)-5588 [99]), (F) S. cerevisiae (EMD-2634) and Homo sapiens (EMD-2635) [57] Mediator complex, (G) S. cerevisiae RNA Pol II–core Mediator transcription initiation complex (EMD-2786 [48]), (H) S. pombe core Mediator (PDB 5N9J [47]) and (I) Homo sapiens MED23 subunit (PDB 6H02 [72]). For details, refer to the text. For a complete collection of structural data on Mediator, see Supplementary Table S1 adapted from [28] with permission from Elsevier. Figures were prepared with PyMol [116] or UCSF Chimera [117]. y, yeast; h, human; m, murine; Sc, Saccharomyces cerevisiae, Sp, Schizosaccharomyces pombe.
Structural studies of Mediator
The modular and dynamic nature of Mediator was pointed out from initial low-resolution EM reconstructions of endogenous complexes [22,25]. Early biochemical and genetic studies revealed many key subunit–subunit interactions that were instrumental to guide initial structural studies [31–41]. During the 2000s, high-resolution structures of yeast Mediator became available for the Med7C/Med21 heterodimer [42] (Figure 1A), the Med8C/Med18/Med20 subcomplex [43,44] (Figure 1B), the Med7N/Med31 submodule [45] and the Med11/Med22 four-helix bundle domain [46] (reviewed in [28]). These structures, although important, were limited to small parts of the whole complex. More recently, developments in the structural analysis of Mediator made high-resolution structural information of large parts of the entire complex a reality [47–50].
Head module
The Mediator Head module is composed of seven highly conserved subunits (Med6, Med8, Med11, Med17, Med18, Med20 and Med22) and together with the Middle module, it plays an essential role during the PIC assembly, contacting the RNA Pol II and stabilizing its interaction with the general transcription factors (GTFs) (reviewed in [18]). The first major breakthrough in the structural characterization of the Head module occurred in 2006 with the successful production in insect cells of a recombinant 7-subunit complex from Saccharomyces cerevisiae [51] that enabled a first negative stain EM analysis [52] and, 5 years later, the determination of its crystal structure at 4.3 Å resolution [53]. The Head module structure has a characteristic shape reminiscent of a wrench constituted by three major domains that were initially named neck, fixed and movable jaws [51–53] (Figure 1C). These features were later confirmed by the structure of the S. cerevisiae Head module in complex with a 35-residues peptide containing five CTD heptad repeats [54] and by the crystal structure of the related S. pombe Head module at 3.4 Å resolution [55] (Figure 1D). This later structure led to a revised and more complete architecture of the Head, which has been described overall by eight distinct elements [55] (Supplementary Figure S1). One remarkable feature of the Head structure is the neck domain that stabilizes the module by forming a multi-helical bundle involving five different subunits, Med6, Med8, Med11, Med17 and Med22 [53–55] (Figure 1C). Importantly, these studies underline the fact that, when expressed individually, recombinant Mediator subunits are generally insoluble and stress the strict requirement for co-expression of subunits constituting a module or a submodule [31,42–46,51–53,56] (reviewed in [28]).
The structure determination of the Mediator Head module [53–55] represented a first major accomplishment towards the characterization of the entire Mediator complex at high resolution. In this context, information about the exact subunit localization and boundaries of the full three main modules was mandatory but remained rather elusive, even contradictory, more than 10 years after their first identification. Indeed at that time, the quality of the available EM structures was limited, as well as the capacity to analyze Mediator samples displaying considerable compositional and/or conformational heterogeneity. In 2014, two pivotal studies [57,58] reported cryo-electron microscopy (cryo-EM) analyses that completely redefined the modular organization of Mediator. Until then the Head module was assigned to one end of the Mediator structure, with the Middle and Tail modules folded one over the other to form the opposite portion of Mediator (reviewed in [59]). Using either tagged or deleted individual subunits combined with unequivocal docking of the Head module X-ray structure, these two studies delivered an improved cryo-EM reconstruction of yeast Mediator at 20–40 Å resolution [57,58] (Figure 1F) that showed, in a dramatic topsy-turvy twist, that the Head and Middle modules were forming the portion of Mediator previously attributed to the Middle and Tail modules, while the large opposite domain corresponded to the Tail, and not to the Head. As a consequence of this revised organization, EM structures of Mediator reported before 2014 should be interpreted with caution. A low-resolution map of human Mediator complex (Figure 1F) was also obtained [57], confirming the overall structural similarity of yeast and human Mediator despite their evolutionary divergence [24]. Previously unassigned metazoan subunits were also clearly localized. For example, Med27, Med28, Med29 and Med30 make extensive contacts with the Head module while Med26 associates with the Middle module [57]. Finally, a recombinant human 8-subunit Head module that additionally contained the metazoan subunit Med30 was successfully reconstituted in insect cells in 2014 [60]. The reconstitution of the recombinant human Head module was done in the context of an assembly of a functional 15-subunit human core Mediator complex that comprises the Head and Middle modules held together by the Med14 subunit (see below) [60]. However, structural data regarding the human Head module are still missing.
Middle module
The Middle module comprises up to nine subunits (Med1, Med4, Med7, Med9, Med10, Med19, Med21 and Med31 as well as Med26 in mammals). The structure of the Middle module remained unknown for a long time and detailed structural information in yeast was limited until 2017 to two small subcomplexes, Med7N/Med31 [45] and Med7C/Med21 [42] (Figure 1A). The heterologous co-expression strategy was again successfully used in 2010 by the Cramer laboratory to purify a recombinant yeast 7-subunit Middle module [61]. In accordance with a previous report [38], an endogenous 7-subunit Middle module had been purified from a Δmed19 strain, and Med19 was thus not included for the subsequent expression of this recombinant Mediator Middle module [61]. Limited proteolysis and co-expression pull-down experiments [61] together with previously published data [32–34,42,62], established at that time the most detailed protein interaction map of the Middle module. However, the high intrinsic flexibility of the module prevented its crystallization. Three years later, bacterial co-expression of a 6-subunit Middle module lacking Med1 combined with cross-linking experiments and homology models, allowed to propose a three-dimensional model of the Middle module [63]. In parallel, reconstitution in insect cells of a human recombinant Mediator 5-subunit Middle module (Med4, Med7, Med10, Med21 and Med31) was also obtained in 2014 [60]. Again, like for the human Head module, no structural data have yet emerged from these studies for the human Middle module. The only known structure of a human Mediator Middle module subunit is the N-terminal domain of Med26 [64] that serves in mammals as an overlapping docking site for super-elongation complexes (SECs) containing ELL/EAF family members as well as TFIID [65].
Core Mediator
Core Mediator, a term that in the past has been used in different contexts, refers now to the minimal set of recombinant Mediator subunits active in transcription, which corresponds to the Head and Middle modules [48,60]. Indeed with the high-resolution structure of the Head module [53–55] and the three-dimensional architectural model of a 6-subunit Middle module [63] at hand in 2013, the next challenge was to decipher their spatial arrangement at the atomic level. But to do so, it appeared that one central piece of the puzzle was still missing. Med14 is one of the largest Mediator subunits and was assigned initially to either the Tail or the Middle module [24,33,38–40]. However, biochemical analyses of Mediator subassemblies in yeast have revealed very early on [35,36,40] that Med14 was tightly associated not only with Tail subunits (Med2, Med3, Med15, Med16) but also with many subunits of the Middle module (Med1, Med4, Med7, Med9 and Med21), suggesting that Med14 was possibly linking different modules. The role of Med14 as a key Mediator scaffold protein was confirmed in 2014 when the improved cryo-EM reconstruction of yeast Mediator at a resolution of 18 Å came out [57,58] (Figure 1F). In particular, C-terminal MBP tagging and N-terminal antibody labeling of Med14 [57] coupled with cross-linking and mass spectrometry of native Mediator [66] clearly indicated that the Med14 N and C termini were located at opposite ends of a large central density spanning ∼220 Å and connecting all three modules. Importantly, the scaffold function of yeast Med14 appeared to be conserved in the human Mediator complex [60].
The Head–Middle module interface, the precise localization of Med14 and the individual Middle module subunits were revealed soon after when two cryo-EM structures were reported, that of S. cerevisiae core Mediator bound to a core initiation complex (cITC–cMed that contains 31 polypeptides) at 9.7 Å resolution [48] (Figure 1G) and that of S. pombe Mediator at 4.4 Å resolution [50] (reviewed in [26,27,29]). The resulting architectural model also revealed the precise location of Mediator on RNA Pol II (see below). Finally, in a long-term effort to solve the high-resolution Mediator structure, the Cramer laboratory reported in 2017 the crystal structure of a 15-subunit core Mediator (cMed) from S. pombe at an impressive 3.4 Å resolution [47] (Figures 1H, 2 and Supplementary Figure S1). The cMed structure is divided into 13 submodules, 8 in the Head as previously defined [55] and 5 in the Middle module (Supplementary Figure S1). Especially noteworthy are the tether regions formed by the C-terminus of Med6 and the N-terminus of Med17 that both associate with Med14 linking the Head and Middle modules [47] and the key architectural roles performed by Med14 and Med17 that make an extensive network of intersubunit interactions, with Med14 connecting all three modules [47,48,50,60,66]. Thus, these structures represent a major advance in our understanding of the structure of yeast core Mediator and overall, only Med1 remains unresolved [47,50].
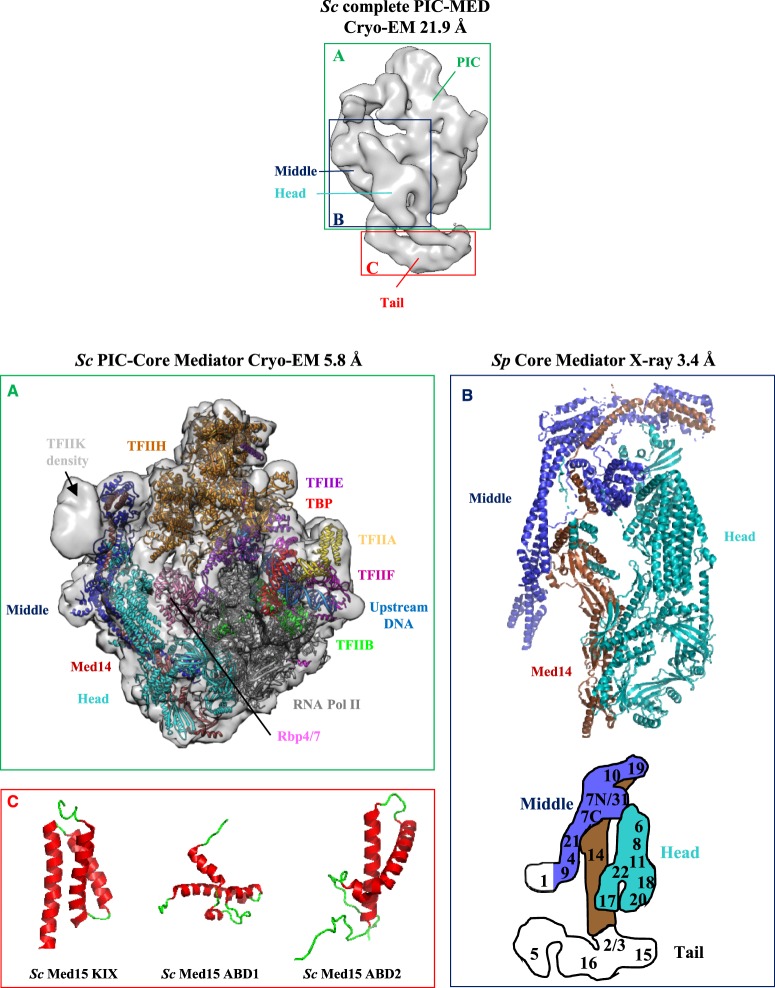
Figure 2. Structural studies of yeast Mediator complex.
Cryo-EM three-dimensional reconstruction of a complete S. cerevisiae Mediator–RNA Pol II preinitiation complex (Sc PIC-MED) at 21.9 Å resolution (EMD-8308 [69]). PIC, Mediator Head, Middle and Tail modules are indicated. Individual panels display high-resolution structures of ((A), in green) S. cerevisiae PIC-core Mediator at 5.8 Å resolution (EMD-3850 [49]) in semi-transparent rendering with the docked atomic model (PDB 5OQM) in ribbon representation. The cryo-EM density for the mobile Kin28–Ccl1 kinase complex of TFIIH (TFIIK) is indicated. ((B), in dark blue) S. pombe core Mediator structure at 3.4 Å resolution (PDB 5N9J [47]). For the sake of simplicity, the Mediator Head module is depicted in cyan and the Middle module in blue, with their respective subunits displayed in ribbon representation. The Med14 subunit is in brown. For a more detailed description of core Mediator structure and its 13 defined submodules, see Supplementary Figure S1. The localization of Mediator subunits is schematized below in the Mediator diagram adapted from [50,57] with the same color code (Head module in cyan, Middle module in blue and Tail module in white). The Middle module Med1 subunit was included in recombinant core Mediator but is lacking in the crystal structure [47]. Med1 is thus colored in white. ((C), in red) NMR structures of S. cerevisiae Med15 KIX domain (PDB 2K0N [77]), Med15 ABD1 domain (PDB 2LPB [74]) and Med15 ABD2 domain (PDB 6ALY [78]).
Tail module
The Tail module subunits are the most evolutionarily divergent [9] and include in S. cerevisiae Med2, Med3, Med5, Med15 and Med16. In addition to the conserved Med15 and Med16, Mediator Tail includes in metazoans three other subunits, Med23, Med24 and Med25 with Med24 considered to be a highly divergent ortholog of yeast Med5 [9] (reviewed in [67]). It has also been suggested that metazoans Med27 and Med29 are very distant orthologs of yeast Med3 and Med2, respectively [9]. These subunits are thus now often referred to in the literature as Med2/29, Med3/27 and Med5/24. Their precise assignment along with the two other metazoan subunits Med28 and Med30 remains to be assessed. Notably, Med27, Med28, Med29 and Med30 make extensive contacts with the Head module [57,60,68] but in S. pombe there is also a clear Med27-Tail connection [50]. As mentioned above, the Med14 subunit makes extensive contacts with all three modules [47–50,57,60,66] and is now viewed as a key architectural backbone of the Mediator complex rather than simply a Tail and/or a Middle subunit.
Notwithstanding, the Tail module remains currently unresolved likely due to its conformational heterogeneity (reviewed in [26]). For example, in a small subset of the S. cerevisiae Mediator–PIC particles [69] (Figure 2), alternate Tail module conformations were observed consistent with the previous observation that the presence of the yeast transcription factor Gcn4 causes a rotation of the Tail module [57]. These conformational changes triggered by transcription factors (within the limits of the very early low-resolution EM reconstructions) are also evident in the human Mediator complex [25,70,71]. Such structural transitions, which seem to propagate throughout the entire Mediator complex, have been hypothesized to promote its stable association with the transcription machinery (reviewed in [12]). These observations raise the possibility that transcription factors capable of coordinating changes in Mediator conformation required for its interaction with RNA Pol II could play major roles in governing transcription. An intriguing possibility is that the Tail module may exert an inhibitory effect on the Head and/or Middle modules in order to prevent promiscuous transcription in absence of activators. Anyhow, until 2018 when the first structure of a whole Mediator Tail subunit (human Med23) was finally solved [72], only small portions of subunits from the Tail module were structurally characterized [73–80] (Figures 1I, 2 and 3). Despite this limited structural information, the overall architecture and subunits interactions are now fairly well characterized, at least in yeast [57,66]. Consistent with early observations that a C-terminal truncation of Med14 caused the loss of Tail subunits [40] and that Med2/Med3/Med15 form a stable subcomplex [31,81], Med2, Med3, Med5, Med15 and Med16 form the bottom domain of the structure and are interconnected to the rest of Mediator through the C-terminus of Med14 [50,57,66]. Importantly, this structural architecture appears to be conserved in the human Mediator complex where Med14 is able to bridge the Head, Middle and Tail modules [60]. However, the human Tail module constitutes a very complex interaction hub and low-resolution Tail EM density of human Mediator at 30 Å resolution appeared much larger that its yeast counterpart [57] (Figure 3). Although the precise metazoan Tail subunit organization has yet to be determined, Med16, Med23 and Med24 are known to form a submodule [82,83]. The reconstitution of a 15-subunit human core Mediator complex [60] and the recent crystal structure of human Med23 [72] are elements that should progressively pave the way for future reconstruction of the full complex architecture.
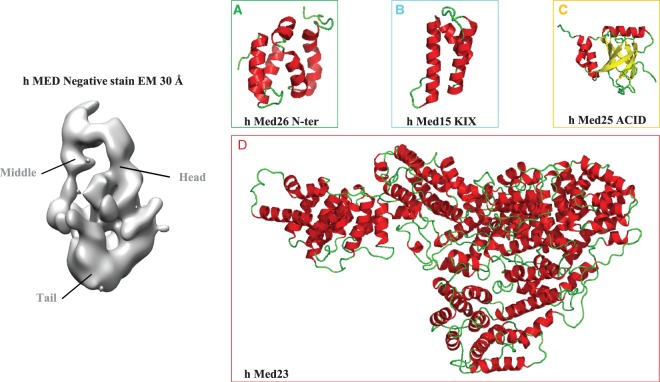
Figure 3. Structural studies of human Mediator complex.
EM map of Human Mediator at 30 Å resolution (EMD-2635 [57]). Mediator Head, Middle and Tail modules are indicated. Individual panels display high-resolution structures of ((A), in green) the N-terminal domain of Human Med26 (PDB 5ODD [64]), ((B), in cyan) KIX domain of human Med15 (PDB 2GUT [80]), ((C), in yellow) ACID/PTOV domain of human Med25 (PDB 2L23, 2XNF, 2L6U and 2KY6 [73,75,76,79]) and ((D), in red) human Med23 (PDB 6H02 [72]). Med26 associates with the Middle domain via interactions with Med4, Med7 and Med19 [57]. Note that Med26 N- and C-terminal domains support interactions with TFIID and transcription elongation factors and with Mediator complex, respectively [65]. Med15 and Med23 have been assigned to the Tail module and Med25, which engages in an extensive network of interactions with other Tail subunits is believed to be a component of the Tail [57]. For details, refer to the text.
Finally, several studies identified Mediator subunits that are contacted by transcription factors and these interactions tend to cluster in the Tail module (reviewed in [13]). Structural analysis using mainly nuclear magnetic resonance (NMR) has revealed the molecular details of interactions between transactivation domains (TAD) of transcription factors and the activator-binding domains (ABD) of a few Mediator Tail subunits including sterol regulatory element-binding protein (SREBP)/Med15 [80], VP16/Med25 [73,76,79], the PEA3 subfamily of Ets transcription factors/Med25 [84,85], ATF6α/Med25 [86] and Gcn4/Med15 [74,78] (Figures 2 and 3). These studies illustrate the conformational heterogeneity of intrinsically unstructured TADs that are engaged in multiple low-affinity and low-specificity interactions with their targets (reviewed in [87]). These multivalent interactions may drive phase-separated condensates that are potentially important for transcriptional control [88–91]. These structural studies are also particularly relevant for human diseases (reviewed in [92]) and the development of small molecules to interfere with transcription factor–Mediator subunit interactions (reviewed in [93]).
CDK8 kinase module
A major point of agreement established long ago is that Mediator subunits are organized into three modules (Head, Middle, Tail) and a four-subunit (CDK8, CycC, Med12 and Med13) dissociable CDK8 kinase module (CKM) [94]. The kinase module has been associated primarily with repressive functions on Mediator but has also been implicated in activation of transcription (reviewed in [19]). Multiple studies have indicated that the RNA Pol II and the CKM interact with the Mediator complex in a mutually exclusive manner [95–99] and that ejection of the kinase module is required for Mediator to join the PIC (reviewed in [27]). The structure of S. pombe CycC [100] and of human CDK8/CycC [101] have been solved but limited information is available for Med12 or Med13. The overall molecular architecture of budding yeast CKM has been obtained in 2013 by cryo-EM at ∼15 Å resolution [99,102] (Figure 1E). Collectively, these two studies demonstrated that Med13 and the CDK8/CycC pair are at opposite end of the CKM structure, connected by Med12 that forms the central lobe [99,102]. Interestingly, the interaction between Mediator and CKM appears very similar in yeast and human [96] Mediators, with the CKM tethered via its Med13 subunit to the hook domain formed by Middle module subunits at the very end of Mediator structure (reviewed in [26,27]). An extended interface with Mediator that overlaps with the position that would be occupied by the RNA pol II holoenzyme was also observed in yeast [95,99], suggesting that Mediator and CKM may interact in different ways. Structural data with human Mediator complex rather suggest that CKM-induced structural rearrangements block Mediator–RNA Pol II interaction, or vice versa [96,103]. The structural basis of this CKM inhibitory interaction awaits a high-resolution structure of the entire Mediator complex with its kinase module.
Structural studies of RNA Pol II–Mediator complexes
Because RNA Pol II and Mediator lie at the center of the transcription machinery, several studies have aimed to characterize the structure of the Mediator–RNA Pol II assembly [22–24,48–50,52,57,58,69,99,103–105] (Supplementary Table S1). Initial EM studies led to inconsistent locations of Mediator on RNA Pol II, probably owing to Mediator heterogeneity and low-resolution reconstruction. Mediator density in yeast was observed at four different locations on RNA Pol II [23,53,54,99,104–106] and a fifth position was obtained in the human RNA Pol II–TFIIF–VP16–Mediator complex [103]. Recent low- [69] and high-resolution cryo-EM structures of S. cerevisiae [48,49] (Figure 2) and S. pombe holoenzymes [50] have been reported and show similar overall structures with subtle differences in detail, resolving previous ambiguities. In these structures, Med18 and Med20 contact the RNA Pol II Rpb3–Rpb11 heterodimer and the TFIIB β-ribbon, Med22 and Med8 contact Rpb4 and Med9 contacts the RNA Pol II foot region [49]. The structural comparison of both unbound Mediator [47] and cITC–cMed [48] with the latest available PIC–cMed cryo-EM structures [49,50,107] suggests a significant repositioning of the Middle module with respect to the Head module upon PIC binding. In particular, four submodules undergo concerted movements owing to large conformational changes in Med14 and inherent structural flexibility of the Med7/Med21 hinge (Supplementary Figure S1). Collectively, these Mediator–Pol II contacts are globally consistent with functional data (reviewed in [18]) but do not exclude the possibility that other conformations could exist during the assembly of the PIC in vivo [108,109].
These extensive core Mediator–RNA Pol II contacts may appear paradoxical given that phosphorylation of the unstructured Pol II CTD is known to drive dissociation of Mediator from RNA Pol II [110–113]. However, to date, it remains unclear how the destabilization of the complex occurs, especially since the path of the CTD along Mediator is still under debate [48–50,54,69]. The recent PIC–cMed cryo-EM structures [48,49] showed evidence that the linker to the mobile CTD extends from RNA Pol II towards the inner surface of a ‘cradle’ formed between RNA Pol II and Mediator to reach the TFIIH kinase (reviewed in [26]). An intriguing possibility is that the global structural organization of Mediator defines the topological and mobility restraints of the entire PIC assembly to properly orient the RNA Pol II CTD for efficient phosphorylation by TFIIH. Phosphorylation of CTD would result in the accumulation of negative charges and thus lead to dissociation of Mediator [49].
Conclusion and perspectives
From the averaging of 54 individual particles in negative stain in 1999 to the highest resolution structures of core Mediator alone and within the PIC determined in 2017 by modern cryo-EM single particle reconstruction techniques, nearly 20 years have passed. During this period, the revolution of integrated structural biology irrigated the studies devoted to the Mediator and allowed unprecedented progress in our quest to reveal its structure that could not have been anticipated or even dreamed ∼10 years ago. While the structures of the Head and Middle modules have been revealed, little structural information is still available for the Tail and CKM and their high-resolution structural determination constitutes one of the next challenges. In addition, the structural impact of the association of the PIC with Mediator and transcription factors remains elusive. It seems now possible, within the next decade, to reveal the structure of the full Mediator complex, both in yeast and human. In a mid-term effort, it should be possible to directly visualize how transcription factors influence the Mediator Tail module structure, how the regulatory signals are transmitted to the core Mediator and subsequently to the PIC, and how the kinase module interferes with Pol II binding. Finally, near-atomic resolution cryo-EM maps of the PIC complex including TFIIH and the Mediator Head and Middle modules already represent major breakthroughs towards these goals and together with the very recent cryo-EM structures of yeast [114] and human [115] TFIID, provide an exciting framework which paves the way for the long awaited structural characterization of the entire initiation machinery.
Acknowledgements
We apologize to those authors whose work could not be cited owing to space restrictions. We are grateful to Zoé Lens, Henri-Marc Bourbon and Muriel Boube for their critical reading of the manuscript. We thank Patrick Cramer for the permission to reuse his 2012 published collection of structural data on Mediator.
Abbreviations
- ACID/PTOV
activator-interacting domain/prostate overexpressed
- CDK8
cyclin-dependent kinase 8
- cITC
core initiation complex
- CKM
CDK8 kinase module
- cMed
core Mediator
- cryo-EM
cryogenic electron microscopy
- CTD
carboxy-terminal domain
- EM
electron microscopy
- GTF
general transcription factor
- KIX
kinase-inducible domain interacting domain
- MED
Mediator
- PIC
PreInitiation complex
- RNA Pol II
RNA polymerase II
- Srb
suppressor of RNA polymerase B
- TAD
transactivation domain
Funding
The authors’ research is supported by CNRS and the French National Research Agency (Grant ANR-DS0401-2016 MEDNET).
Author Contribution
A.V. wrote the first draft. A.V., D.M. and V.V. edited and approved the final version of the manuscript.
Competing Interests
The Authors declare that there are no competing interests associated with the manuscript.
Supplementary Material
References
- 1.Kelleher, R.J. III, Flanagan P.M. and Kornberg R.D. (1990) A novel mediator between activator proteins and the RNA polymerase II transcription apparatus. Cell 61, 1209–1215 10.1016/0092-8674(90)90685-8 [DOI] [PubMed] [Google Scholar]
- 2.Flanagan P.M., Kelleher, R.J. III, Sayre M.H., Tschochner H. and Kornberg R.D. (1991) A mediator required for activation of RNA polymerase II transcription in vitro. Nature 350, 436–438 10.1038/350436a0 [DOI] [PubMed] [Google Scholar]
- 3.Nonet M.L. and Young R.A. (1989) Intragenic and extragenic suppressors of mutations in the heptapeptide repeat domain of Saccharomyces cerevisiae RNA polymerase II. Genetics 123, 715–724 PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thompson C.M., Koleske A.J., Chao D.M. and Young R.A. (1993) A multisubunit complex associated with the RNA polymerase II CTD and TATA-binding protein in yeast. Cell 73, 1361–1375 10.1016/0092-8674(93)90362-T [DOI] [PubMed] [Google Scholar]
- 5.Kim Y.J., Björklund S., Li Y., Sayre M.H. and Kornberg R.D. (1994) A multiprotein mediator of transcriptional activation and its interaction with the C-terminal repeat domain of RNA polymerase II. Cell 77, 599–608 10.1016/0092-8674(94)90221-6 [DOI] [PubMed] [Google Scholar]
- 6.Myers L.C. and Kornberg R.D. (2000) Mediator of transcriptional regulation. Annu. Rev. Biochem. 69, 729–749 10.1146/annurev.biochem.69.1.729 [DOI] [PubMed] [Google Scholar]
- 7.Conaway R.C., Sato S., Tomomori-Sato C., Yao T. and Conaway J.W. (2005) The mammalian mediator complex and its role in transcriptional regulation. Trends Biochem. Sci. 30, 250–255 10.1016/j.tibs.2005.03.002 [DOI] [PubMed] [Google Scholar]
- 8.Boube M., Joulia L., Cribbs D.L. and Bourbon H.M. (2002) Evidence for a mediator of RNA polymerase II transcriptional regulation conserved from yeast to man. Cell 110, 143–151 10.1016/S0092-8674(02)00830-9 [DOI] [PubMed] [Google Scholar]
- 9.Bourbon H.M. (2008) Comparative genomics supports a deep evolutionary origin for the large, four-module transcriptional mediator complex. Nucleic Acids Res. 36, 3993–4008 10.1093/nar/gkn349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sato S., Tomomori-Sato C., Parmely T.J., Florens L., Zybailov B., Swanson S.K. et al. (2004) A set of consensus mammalian mediator subunits identified by multidimensional protein identification technology. Mol. Cell 14, 685–691 10.1016/j.molcel.2004.05.006 [DOI] [PubMed] [Google Scholar]
- 11.Bourbon H.M., Aguilera A., Ansari A.Z., Asturias F.J., Berk A.J., Björklund S. et al. (2004) A unified nomenclature for protein subunits of mediator complexes linking transcriptional regulators to RNA polymerase II. Mol. Cell 14, 553–557 10.1016/j.molcel.2004.05.011 [DOI] [PubMed] [Google Scholar]
- 12.Allen B.L. and Taatjes D.J. (2015) The mediator complex: a central integrator of transcription. Nat. Rev. Mol. Cell Biol. 16, 155–166 10.1038/nrm3951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Borggrefe T. and Yue X. (2011) Interactions between subunits of the mediator complex with gene-specific transcription factors. Semin. Cell Dev. Biol. 22, 759–768 10.1016/j.semcdb.2011.07.022 [DOI] [PubMed] [Google Scholar]
- 14.Conaway R.C. and Conaway J.W. (2011) Function and regulation of the mediator complex. Curr. Opin. Genet. Dev. 21, 225–230 10.1016/j.gde.2011.01.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Conaway R.C. and Conaway J.W. (2013) The mediator complex and transcription elongation. Biochim. Biophys. Acta 1829, 69–75 10.1016/j.bbagrm.2012.08.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Malik S. and Roeder R.G. (2010) The metazoan mediator co-activator complex as an integrative hub for transcriptional regulation. Nat. Rev. Genet. 11, 761–772 10.1038/nrg2901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Poss Z.C., Ebmeier C.C. and Taatjes D.J. (2013) The mediator complex and transcription regulation. Crit. Rev. Biochem. Mol. Biol. 48, 575–608 10.3109/10409238.2013.840259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Soutourina J. (2018) Transcription regulation by the mediator complex. Nat. Rev. Mol. Cell Biol. 19, 262–274 10.1038/nrm.2017.115 [DOI] [PubMed] [Google Scholar]
- 19.Yin J.W. and Wang G. (2014) The mediator complex: a master coordinator of transcription and cell lineage development. Development 141, 977–987 10.1242/dev.098392 [DOI] [PubMed] [Google Scholar]
- 20.Nagulapalli M., Maji S., Dwivedi N., Dahiya P. and Thakur J.K. (2016) Evolution of disorder in mediator complex and its functional relevance. Nucleic Acids Res. 44, 1591–1612 10.1093/nar/gkv1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tóth-Petróczy A., Oldfield C.J., Simon I., Takagi Y., Dunker A.K., Uversky V.N. et al. (2008) Malleable machines in transcription regulation: the mediator complex. PLoS Comput. Biol. 4, e1000243 10.1371/journal.pcbi.1000243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Asturias F.J., Jiang Y.W., Myers L.C., Gustafsson C.M. and Kornberg R.D. (1999) Conserved structures of mediator and RNA polymerase II holoenzyme. Science 283, 985–987 10.1126/science.283.5404.985 [DOI] [PubMed] [Google Scholar]
- 23.Davis J.A., Takagi Y., Kornberg R.D. and Asturias F.A. (2002) Structure of the yeast RNA polymerase II holoenzyme: mediator conformation and polymerase interaction. Mol. Cell 10, 409–415 10.1016/S1097-2765(02)00598-1 [DOI] [PubMed] [Google Scholar]
- 24.Dotson M.R., Yuan C.X., Roeder R.G., Myers L.C., Gustafsson C.M., Jiang Y.W. et al. (2000) Structural organization of yeast and mammalian mediator complexes. Proc. Natl Acad. Sci. U.S.A. 97, 14307–14310 10.1073/pnas.260489497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Taatjes D.J., Näär A.M., Andel, F. III, Nogales E. and Tjian R. (2002) Structure, function, and activator-induced conformations of the CRSP coactivator. Science 295, 1058–1062 10.1126/science.1065249 [DOI] [PubMed] [Google Scholar]
- 26.Harper T.M. and Taatjes D.J. (2018) The complex structure and function of mediator. J. Biol. Chem. 293, 13778–13785 10.1074/jbc.R117.794438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jeronimo C. and Robert F. (2017) The mediator complex: at the nexus of RNA polymerase II transcription. Trends Cell Biol. 27, 765–783 10.1016/j.tcb.2017.07.001 [DOI] [PubMed] [Google Scholar]
- 28.Larivière L., Seizl M. and Cramer P. (2012) A structural perspective on mediator function. Curr. Opin. Cell Biol. 24, 305–313 10.1016/j.ceb.2012.01.007 [DOI] [PubMed] [Google Scholar]
- 29.Plaschka C., Nozawa K. and Cramer P. (2016) mediator architecture and RNA polymerase II interaction. J. Mol. Biol. 428, 2569–2574 10.1016/j.jmb.2016.01.028 [DOI] [PubMed] [Google Scholar]
- 30.Hanske J., Sadian Y. and Müller C.W. (2018) The cryo-EM resolution revolution and transcription complexes. Curr. Opin. Struct. Biol. 52, 8–15 10.1016/j.sbi.2018.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Béve J., Hu G.Z., Myers L.C., Balciunas D., Werngren O., Hultenby K. et al. (2005) The structural and functional role of Med5 in the yeast mediator tail module. J. Biol. Chem. 280, 41366–41372 10.1074/jbc.M511181200 [DOI] [PubMed] [Google Scholar]
- 32.Gromoller A. and Lehming N. (2000) Srb7p is a physical and physiological target of Tup1p. EMBO J. 19, 6845–6852 10.1093/emboj/19.24.6845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guglielmi B., van Berkum N.L., Klapholz B., Bijma T., Boube M., Boschiero C. et al. (2004) A high resolution protein interaction map of the yeast mediator complex. Nucleic Acids Res. 32, 5379–5391 10.1093/nar/gkh878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kang J.S., Kim S.H., Hwang M.S., Han S.J., Lee Y.C. and Kim Y.J. (2001) The structural and functional organization of the yeast mediator complex. J. Biol. Chem. 276, 42003–42010 10.1074/jbc.M105961200 [DOI] [PubMed] [Google Scholar]
- 35.Lee Y.C., Park J.M., Min S., Han S.J. and Kim Y.J. (1999) An activator binding module of yeast RNA polymerase II holoenzyme. Mol. Cell Biol. 19, 2967–2976 10.1128/MCB.19.4.2967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee Y.C. and Kim Y.J. (1998) Requirement for a functional interaction between mediator components Med6 and Srb4 in RNA polymerase II transcription. Mol. Cell Biol. 18, 5364–5370 10.1128/MCB.18.9.5364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Koh S.S., Ansari A.Z., Ptashne M. and Young R.A. (1998) An activator target in the RNA polymerase II holoenzyme. Mol. Cell 1, 895–904 10.1016/S1097-2765(00)80088-X [DOI] [PubMed] [Google Scholar]
- 38.Baidoobonso S.M., Guidi B.W. and Myers L.C. (2007) Med19(Rox3) regulates intermodule interactions in the Saccharomyces cerevisiae mediator complex. J. Biol. Chem. 282, 5551–5559 10.1074/jbc.M609484200 [DOI] [PubMed] [Google Scholar]
- 39.Jiang Y.W., Dohrmann P.R. and Stillman D.J. (1995) Genetic and physical interactions between yeast RGR1 and SIN4 in chromatin organization and transcriptional regulation. Genetics 140, 47–54 PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li Y., Bjorklund S., Jiang Y.W., Kim Y.J., Lane W.S., Stillman D.J. et al. (1995) Yeast global transcriptional regulators Sin4 and Rgr1 are components of mediator complex/RNA polymerase II holoenzyme. Proc. Natl Acad. Sci. U.S.A. 92, 10864–10868 10.1073/pnas.92.24.10864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu Y., Ranish J.A., Aebersold R. and Hahn S. (2001) Yeast nuclear extract contains two major forms of RNA polymerase II mediator complexes. J. Biol. Chem. 276, 7169–7175 10.1074/jbc.M009586200 [DOI] [PubMed] [Google Scholar]
- 42.Baumli S., Hoeppner S. and Cramer P. (2005) A conserved mediator hinge revealed in the structure of the MED7.MED21 (Med7.Srb7) heterodimer. J. Biol. Chem. 280, 18171–18178 10.1074/jbc.M413466200 [DOI] [PubMed] [Google Scholar]
- 43.Larivière L., Geiger S., Hoeppner S., Röther S., Sträßer K. and Cramer P. (2006) Structure and TBP binding of the mediator head subcomplex Med8-Med18-Med20. Nat. Struct. Mol. Biol. 13, 895–901 10.1038/nsmb1143 [DOI] [PubMed] [Google Scholar]
- 44.Larivière L., Seizl M., van Wageningen S., Rother S., van de Pasch L., Feldmann H. et al. (2008) Structure-system correlation identifies a gene regulatory mediator submodule. Genes Dev. 22, 872–877 10.1101/gad.465108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Koschubs T., Seizl M., Larivière L., Kurth F., Baumli S., Martin D.E. et al. (2009) Identification, structure, and functional requirement of the mediator submodule Med7N/31. EMBO J. 28, 69–80 10.1038/emboj.2008.254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Seizl M., Larivière L., Pfaffeneder T., Wenzeck L. and Cramer P. (2011) Mediator head subcomplex Med11/22 contains a common helix bundle building block with a specific function in transcription initiation complex stabilization. Nucleic Acids Res. 39, 6291–6304 10.1093/nar/gkr229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nozawa K., Schneider T.R. and Cramer P. (2017) Core mediator structure at 3.4 Å extends model of transcription initiation complex. Nature 545, 248–251 10.1038/nature22328 [DOI] [PubMed] [Google Scholar]
- 48.Plaschka C., Larivière L., Wenzeck L., Seizl M., Hemann M., Tegunov D. et al. (2015) Architecture of the RNA polymerase II-mediator core initiation complex. Nature 518, 376–380 10.1038/nature14229 [DOI] [PubMed] [Google Scholar]
- 49.Schilbach S., Hantsche M., Tegunov D., Dienemann C., Wigge C., Urlaub H. et al. (2017) Structures of transcription pre-initiation complex with TFIIH and mediator. Nature 551, 204–209 10.1038/nature24282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tsai K.L., Yu X., Gopalan S., Chao T.C., Zhang Y., Florens L. et al. (2017) Mediator structure and rearrangements required for holoenzyme formation. Nature 544, 196–201 10.1038/nature21393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Takagi Y., Calero G., Komori H., Brown J.A., Ehrensberger A.H., Hudmon A. et al. (2006) Head module control of mediator interactions. Mol. Cell 23, 355–364 10.1016/j.molcel.2006.06.007 [DOI] [PubMed] [Google Scholar]
- 52.Cai G., Imasaki T., Yamada K., Cardelli F., Takagi Y. and Asturias F.J. (2010) Mediator head module structure and functional interactions. Nat. Struct. Mol. Biol. 17, 273–279 10.1038/nsmb.1757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Imasaki T., Calero G., Cai G., Tsai K.L., Yamada K., Cardelli F. et al. (2011) Architecture of the mediator head module. Nature 475, 240–243 10.1038/nature10162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Robinson P.J., Bushnell D.A., Trnka M.J., Burlingame A.L. and Kornberg R.D. (2012) Structure of the mediator head module bound to the carboxy-terminal domain of RNA polymerase II. Proc. Natl Acad. Sci. U.S.A. 109, 17931–17935 10.1073/pnas.1215241109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Larivière L., Plaschka C., Seizl M., Wenzeck L., Kurth F. and Cramer P. (2012) Structure of the mediator head module. Nature 492, 448–451 10.1038/nature11670 [DOI] [PubMed] [Google Scholar]
- 56.Shaikhibrahim Z., Rahaman H., Wittung-Stafshede P. and Bjorklund S. (2009) Med8, Med18, and Med20 subunits of the mediator head domain are interdependent upon each other for folding and complex formation. Proc. Natl Acad. Sci. U.S.A. 106, 20728–20733 10.1073/pnas.0907645106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tsai K.L., Tomomori-Sato C., Sato S., Conaway R.C., Conaway J.W. and Asturias F.J. (2014) Subunit architecture and functional modular rearrangements of the transcriptional mediator complex. Cell 157, 1430–1444 10.1016/j.cell.2014.05.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang X., Sun Q., Ding Z., Ji J., Wang J., Kong X. et al. (2014) Redefining the modular organization of the core mediator complex. Cell Res. 24, 796–808 10.1038/cr.2014.64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chadick J.Z. and Asturias F.J. (2005) Structure of eukaryotic mediator complexes. Trends Biochem. Sci. 30, 264–271 10.1016/j.tibs.2005.03.001 [DOI] [PubMed] [Google Scholar]
- 60.Cevher M.A., Shi Y., Li D., Chait B.T., Malik S. and Roeder R.G. (2014) Reconstitution of active human core mediator complex reveals a critical role of the MED14 subunit. Nat. Struct. Mol. Biol. 21, 1028–1034 10.1038/nsmb.2914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Koschubs T., Lorenzen K., Baumli S., Sandström S., Heck A.J. and Cramer P. (2010) Preparation and topology of the mediator middle module. Nucleic Acids Res. 38, 3186–3195 10.1093/nar/gkq029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Takahashi H., Kasahara K. and Kokubo T. (2009) Saccharomyces cerevisiae Med9 comprises two functionally distinct domains that play different roles in transcriptional regulation. Genes Cells 14, 53–67 10.1111/j.1365-2443.2008.01250.x [DOI] [PubMed] [Google Scholar]
- 63.Larivière L., Plaschka C., Seizl M., Petrotchenko E.V., Wenzeck L., Borchers C.H. et al. (2013) Model of the mediator middle module based on protein cross-linking. Nucleic Acids Res. 41, 9266–9273 10.1093/nar/gkt704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lens Z., Cantrelle F.X., Peruzzini R., Hanoulle X., Dewitte F., Ferreira E. et al. (2017) Solution structure of the N-terminal domain of mediator subunit MED26 and molecular characterization of its interaction with EAF1 and TAF7. J. Mol. Biol. 429, 3043–3055 10.1016/j.jmb.2017.09.001 [DOI] [PubMed] [Google Scholar]
- 65.Takahashi H., Parmely T.J., Sato S., Tomomori-Sato C., Banks C.A., Kong S.E. et al. (2011) Human mediator subunit MED26 functions as a docking site for transcription elongation factors. Cell 146, 92–104 10.1016/j.cell.2011.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Robinson P.J., Trnka M.J., Pellarin R., Greenberg C.H., Bushnell D.A., Davis R. et al. (2015) Molecular architecture of the yeast mediator complex. Elife 4, e08719 10.7554/eLife.08719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Conaway R.C. and Conaway J.W. (2011) Origins and activity of the mediator complex. Semin. Cell Dev. Biol. 22, 729–734 10.1016/j.semcdb.2011.07.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sato S., Tomomori-Sato C., Banks C.A., Parmely T.J., Sorokina I., Brower C.S. et al. (2003) A mammalian homolog of Drosophila melanogaster transcriptional coactivator intersex is a subunit of the mammalian mediator complex. J. Biol. Chem. 278, 49671–49674 10.1074/jbc.C300444200 [DOI] [PubMed] [Google Scholar]
- 69.Robinson P.J., Trnka M.J., Bushnell D.A., Davis R.E., Mattei P.J., Burlingame A.L. et al. (2016) Structure of a complete mediator-RNA polymerase II pre-initiation complex. Cell 166, 1411–1422 e1416 10.1016/j.cell.2016.08.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Meyer K.D., Lin S.C., Bernecky C., Gao Y. and Taatjes D.J. (2010) P53 activates transcription by directing structural shifts in mediator. Nat. Struct. Mol. Biol. 17, 753–760 10.1038/nsmb.1816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Taatjes D.J., Schneider-Poetsch T. and Tjian R. (2004) Distinct conformational states of nuclear receptor-bound CRSP-Med complexes. Nat. Struct. Mol. Biol. 11, 664–671 10.1038/nsmb789 [DOI] [PubMed] [Google Scholar]
- 72.Monté D., Clantin B., Dewitte F., Lens Z., Rucktooa P., Pardon E. et al. (2018) Crystal structure of human mediator subunit MED23. Nat. Commun. 9, 3389 10.1038/s41467-018-05967-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bontems F., Verger A., Dewitte F., Lens Z., Baert J.L., Ferreira E. et al. (2011) NMR structure of the human mediator MED25 ACID domain. J. Struct. Biol. 174, 245–251 10.1016/j.jsb.2010.10.011 [DOI] [PubMed] [Google Scholar]
- 74.Brzovic P.S., Heikaus C.C., Kisselev L., Vernon R., Herbig E., Pacheco D. et al. (2011) The acidic transcription activator gcn4 binds the mediator subunit gal11/med15 using a simple protein interface forming a fuzzy complex. Mol. Cell 44, 942–953 10.1016/j.molcel.2011.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Eletsky A., Ruyechan W.T., Xiao R., Acton T.B., Montelione G.T. and Szyperski T. (2011) Solution NMR structure of MED25 (391-543) comprising the activator-interacting domain (ACID) of human mediator subunit 25. J. Struct. Funct. Genomics 12, 159–166 10.1007/s10969-011-9115-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Milbradt A.G., Kulkarni M., Yi T., Takeuchi K., Sun Z.J., Luna R.E. et al. (2011) Structure of the VP16 transactivator target in ARC/mediator. Nat. Struct. Biol. 18, 410–415 10.1038/nsmb.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Thakur J.K., Arthanari H., Yang F., Pan S.J., Fan X., Breger J. et al. (2008) A nuclear receptor-like pathway regulating multidrug resistance in fungi. Nature 452, 604–609 10.1038/nature06836 [DOI] [PubMed] [Google Scholar]
- 78.Tuttle L.M., Pacheco D., Warfield L., Luo J., Ranish J., Hahn S. et al. (2018) Gcn4-mediator specificity is mediated by a large and dynamic fuzzy protein-protein complex. Cell Rep. 22, 3251–3264 10.1016/j.celrep.2018.02.097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Vojnic E., Mourão A., Seizl M., Simon B., Wenzeck L., Larivière L. et al. (2011) The mediator MED25 activator interaction domain: structure and cooperative binding of VP16 subdomains. Nat. Struct. Biol. 18, 404–409 10.1038/nsmb.1997 [DOI] [PubMed] [Google Scholar]
- 80.Yang F., Vought B.W., Satterlee J.S., Walker A.K., Jim Sun Z.Y., Watts J.L. et al. (2006) An ARC/mediator subunit required for SREBP control of cholesterol and lipid homeostasis. Nature 442, 700–704 10.1038/nature04942 [DOI] [PubMed] [Google Scholar]
- 81.Zhang F., Sumibcay L., Hinnebusch A.G. and Swanson M.J. (2004) A triad of subunits from the Gal11/tail domain of Srb mediator is an in vivo target of transcriptional activator Gcn4p. Mol. Cell. Biol. 24, 6871–6886 10.1128/MCB.24.15.6871-6886.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ito M., Okano H.J., Darnell R.B. and Roeder R.G. (2002) The TRAP100 component of the TRAP/mediator complex is essential in broad transcriptional events and development. EMBO J. 21, 3464–3475 10.1093/emboj/cdf348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Stevens J.L., Cantin G.T., Wang G., Shevchenko A., Shevchenko A. and Berk A.J. (2002) Transcription control by E1A and MAP kinase pathway via Sur2 mediator subunit. Science 296, 755–758 10.1126/science.1068943 [DOI] [PubMed] [Google Scholar]
- 84.Currie S.L., Doane J.J., Evans K.S., Bhachech N., Madison B.J., Lau D.K.W. et al. (2017) ETV4 and AP1 transcription factors form multivalent interactions with three sites on the MED25 activator-interacting domain. J. Mol. Biol. 429, 2975–2995 10.1016/j.jmb.2017.06.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Landrieu I., Verger A., Baert J.L., Rucktooa P., Cantrelle F.X., Dewitte F. et al. (2015) Characterization of ERM transactivation domain binding to the ACID/PTOV domain of the mediator subunit MED25. Nucleic Acids Res. 43, 7110–7121 10.1093/nar/gkv650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Henderson A.R., Henley M.J., Foster N.J., Peiffer A.L., Beyersdorf M.S., Stanford K.D. et al. (2018) Conservation of coactivator engagement mechanism enables small-molecule allosteric modulators. Proc. Natl Acad. Sci. U.S.A. 115, 8960–8965 10.1073/pnas.1806202115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Erkine A.M. (2018) ‘Nonlinear’ biochemistry of nucleosome detergents. Trends Biochem. Sci. 43, 951–959 10.1016/j.tibs.2018.09.006 [DOI] [PubMed] [Google Scholar]
- 88.Boehning M., Dugast-Darzacq C., Rankovic M., Hansen A.S., Yu T., Marie-Nelly H. et al. (2018) RNA polymerase II clustering through carboxy-terminal domain phase separation. Nat. Struct. Mol. Biol. 25, 833–840 10.1038/s41594-018-0112-y [DOI] [PubMed] [Google Scholar]
- 89.Boija A., Klein I.A., Sabari B.R., Dall'Agnese A., Coffey E.L., Zamudio A.V. et al. (2018) Transcription factors activate genes through the phase-separation capacity of their activation domains. Cell 175, 1842–1855 e1816 10.1016/j.cell.2018.10.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Cho W.K., Spille J.H., Hecht M., Lee C., Li C., Grube V. et al. (2018) mediator and RNA polymerase II clusters associate in transcription-dependent condensates. Science 361, 412–415 10.1126/science.aar4199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sabari B.R., Dall'Agnese A., Boija A., Klein I.A., Coffey E.L., Shrinivas K. et al. (2018) Coactivator condensation at super-enhancers links phase separation and gene control. Science 361, eaar3958 10.1126/science.aar3958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Spaeth J.M., Kim N.H. and Boyer T.G. (2011) Mediator and human disease. Semin. Cell Dev. Biol. 22, 776–787 10.1016/j.semcdb.2011.07.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Phillips A.J. and Taatjes D.J. (2013) Small molecule probes to target the human mediator complex. Isr. J. Chem. 53, 588–595 10.1002/ijch.201300039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Borggrefe T., Davis R., Erdjument-Bromage H., Tempst P. and Kornberg R.D. (2002) A complex of the Srb8, -9, -10, and -11 transcriptional regulatory proteins from yeast. J. Biol. Chem. 277, 44202–44207 10.1074/jbc.M207195200 [DOI] [PubMed] [Google Scholar]
- 95.Elmlund H., Baraznenok V., Lindahl M., Samuelsen C.O., Koeck P.J., Holmberg S. et al. (2006) The cyclin-dependent kinase 8 module sterically blocks mediator interactions with RNA polymerase II. Proc. Natl Acad. Sci. U.S.A. 103, 15788–15793 10.1073/pnas.0607483103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Knuesel M.T., Meyer K.D., Bernecky C. and Taatjes D.J. (2009) The human CDK8 subcomplex is a molecular switch that controls mediator coactivator function. Genes Dev. 23, 439–451 10.1101/gad.1767009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Myers L.C., Gustafsson C.M., Bushnell D.A., Lui M., Erdjument-Bromage H., Tempst P. et al. (1998) The Med proteins of yeast and their function through the RNA polymerase II carboxy-terminal domain. Genes Dev. 12, 45–54 10.1101/gad.12.1.45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Näär A.M., Taatjes D.J., Zhai W., Nogales E. and Tjian R. (2002) Human CRSP interacts with RNA polymerase II CTD and adopts a specific CTD-bound conformation. Genes Dev. 16, 1339–1344 10.1101/gad.987602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Tsai K.L., Sato S., Tomomori-Sato C., Conaway R.C., Conaway J.W. and Asturias F.J. (2013) A conserved mediator-CDK8 kinase module association regulates mediator-RNA polymerase II interaction. Nat. Struct. Mol. Biol. 20, 611–619 10.1038/nsmb.2549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Hoeppner S., Baumli S. and Cramer P. (2005) Structure of the mediator subunit cyclin C and its implications for CDK8 function. J. Mol. Biol. 350, 833–842 10.1016/j.jmb.2005.05.041 [DOI] [PubMed] [Google Scholar]
- 101.Schneider E.V., Böttcher J., Blaesse M., Neumann L., Huber R. and Maskos K. (2011) The structure of CDK8/CycC implicates specificity in the CDK/Cyclin family and reveals interaction with a deep pocket binder. J. Mol. Biol. 412, 251–266 10.1016/j.jmb.2011.07.020 [DOI] [PubMed] [Google Scholar]
- 102.Wang X., Wang J., Ding Z., Ji J., Sun Q. and Cai G. (2013) Structural flexibility and functional interaction of mediator Cdk8 module. Protein Cell 4, 911–920 10.1007/s13238-013-3069-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Bernecky C., Grob P., Ebmeier C.C., Nogales E. and Taatjes D.J. (2011) Molecular architecture of the human mediator-RNA polymerase II-TFIIF assembly. PLoS Biol. 9, e1000603 10.1371/journal.pbio.1000603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Bushnell D.A. and Kornberg R.D. (2003) Complete, 12-subunit RNA polymerase II at 4.1-A resolution: implications for the initiation of transcription. Proc. Natl Acad. Sci. U.S.A. 100, 6969–6973 10.1073/pnas.1130601100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Cai G., Imasaki T., Takagi Y. and Asturias F.J. (2009) Mediator structural conservation and implications for the regulation mechanism. Structure 17, 559–567 10.1016/j.str.2009.01.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Cai G., Chaban Y.L., Imasaki T., Kovacs J.A., Calero G., Penczek P.A. et al. (2012) Interaction of the mediator head module with RNA polymerase II. Structure 20, 899–910 10.1016/j.str.2012.02.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Sato S., Tomomori-Sato C., Tsai K.L., Yu X., Sardiu M., Saraf A. et al. (2016) Role for the MED21-MED7 hinge in assembly of the mediator-RNA polymerase II holoenzyme. J. Biol. Chem. 291, 26886–26898 10.1074/jbc.M116.756098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Eychenne T., Novikova E., Barrault M.B., Alibert O., Boschiero C., Peixeiro N. et al. (2016) Functional interplay between mediator and TFIIB in preinitiation complex assembly in relation to promoter architecture. Genes Dev. 30, 2119–2132 10.1101/gad.285775.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Soutourina J., Wydau S., Ambroise Y., Boschiero C. and Werner M. (2011) Direct interaction of RNA polymerase II and mediator required for transcription in vivo. Science 331, 1451–1454 10.1126/science.1200188 [DOI] [PubMed] [Google Scholar]
- 110.Jeronimo C. and Robert F. (2014) Kin28 regulates the transient association of mediator with core promoters. Nat. Struct. Mol. Biol. 21, 449–455 10.1038/nsmb.2810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Svejstrup J.Q., Li Y., Fellows J., Gnatt A., Bjorklund S. and Kornberg R.D. (1997) Evidence for a mediator cycle at the initiation of transcription. Proc. Natl Acad. Sci. U.S.A. 94, 6075–6078 10.1073/pnas.94.12.6075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Wong K.H., Jin Y. and Struhl K. (2014) TFIIH phosphorylation of the Pol II CTD stimulates mediator dissociation from the preinitiation complex and promoter escape. Mol. Cell 54, 601–612 10.1016/j.molcel.2014.03.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Søgaard T.M. and Svejstrup J.Q. (2007) Hyperphosphorylation of the C-terminal repeat domain of RNA polymerase II facilitates dissociation of its complex with mediator. J. Biol. Chem. 282, 14113–14120 10.1074/jbc.M701345200 [DOI] [PubMed] [Google Scholar]
- 114.Kolesnikova O., Ben-Shem A., Luo J., Ranish J., Schultz P. and Papai G. (2018) Molecular structure of promoter-bound yeast TFIID. Nat. Commun. 9, 4666 10.1038/s41467-018-07096-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Patel A.B., Louder R.K., Greber B.J., Grünberg S., Luo J., Fang J. et al. (2018) Structure of human TFIID and mechanism of TBP loading onto promoter DNA. Science 362, eaau8872 10.1126/science.aau8872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.PYMOL The PyMOL Molecular Graphics System, Version 2.0.6, Schrödinger, LLC [Google Scholar]
- 117.Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C. et al. (2004) UCSF chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 10.1002/jcc.20084 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.