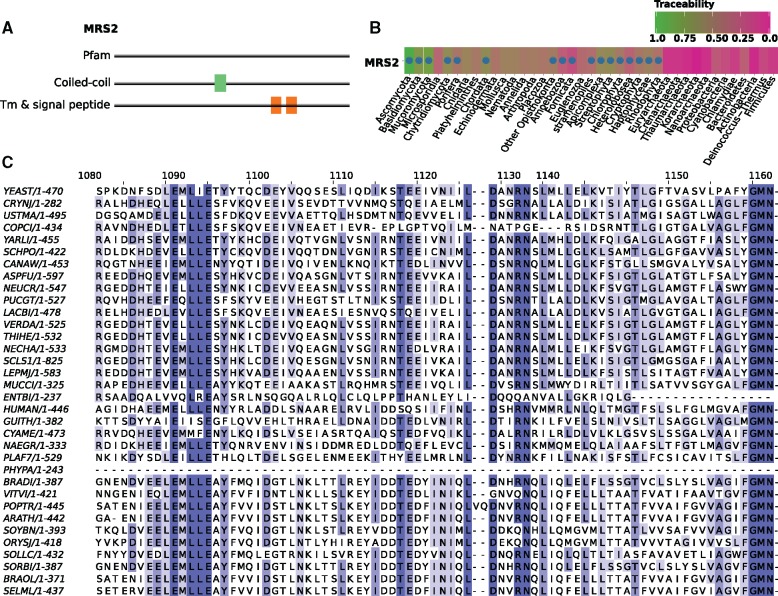
Fig. 3.
—Missing information about domain constraints results in underestimated traceabilities: the yeast mitochondrial inner membrane Mg2+ transporter MRS2. (A) MRS2 displays no significant hit against any Pfam domain and contains as sole features a central coiled-coil domain and two transmembrane domains. (B) The phylogenetic profile of MRS2 reveals the existence of orthologs across the entire eukaryotic kingdoms despite a predicted low traceability. The presence of an ortholog in a given species is indicated by a dot. The cell color represents protein traceability. (C) Section of the MRS2 alignment considering orthologs from different representatives across the eukaryotic tree of life. The selected region shows exemplarily for the entire alignment that MRS2 orthologs share conserved sequence motifs that most likely are associated with the functionality of this protein as an Mg2+ membrane transporter. As these conserved domains are not represented in a Pfam domain, protTrace cannot consider the corresponding evolutionary constraints during its simulation.