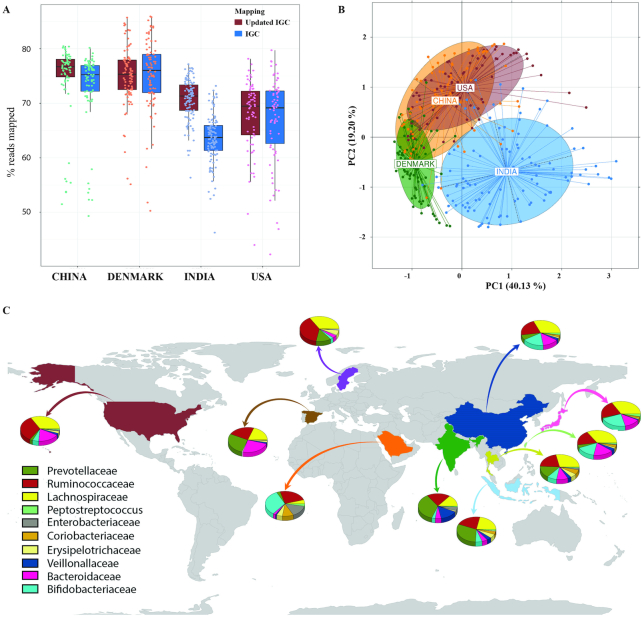
Figure 1:
Comparison of the Indian gut microbiome with other major populations using 16S rRNA gene and metagenomic datasets. (A) Percentage of total reads that could be mapped to IGC and updated IGC containing the Indian gene catalogue. Plotted are interquartile ranges (IQR; in boxes), median (as dark lines in the boxes), lowest and highest values within 1.5 times the IQR (shown as whiskers extending from boxes), and outliers as points beyond these whiskers. The blue and red boxes show percentage of reads mapped to IGC and updated IGC (containing the Indian microbial genes). (B) Principal component analysis using metagenomic species (MGS)/co-abundance gene group (CAG) proportion derived from a metagenome-wide association study. Variations across populations are shown using PC1 and PC2. (C) Illustration of proportions of bacterial families in different populations and their composition as determined from 16S rRNA gene datasets (adult population only). The mean family compositions of abundant families (≥1%) are represented in separate pie plots from 10 different country-wise datasets, showing their overall microbial composition compared to Indian population.