Fig. 1.
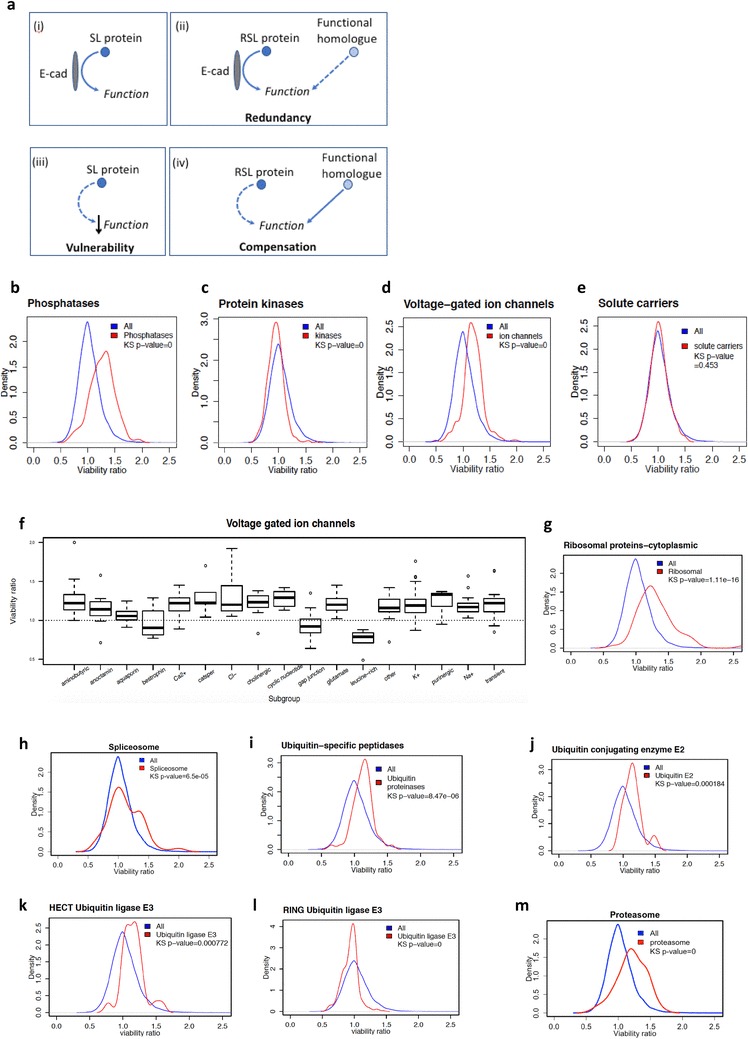
a Model of SL and RSL interactions. SL and RSL proteins both have functions affected by E-cadherin activity; however, RSL proteins have the potential to be compensated for by functional homologues (i–ii). When E-cadherin is lost, inhibition of an SL protein leads to reduced cell fitness (iii), whereas the inhibition of an RSL protein has less effect because its function is compensated for by the increased activity of the functional homologue (iv). b–e Density distributions of MCF10A-CDH1−/−/MCF10A viability ratios following siRNA knockdown of families of phosphatases, protein kinases, voltage-gated ion channels and solute carriers. Lists were obtained from a variety of sources: phosphatases from the human dephosphorylation database (http://www.Koehn.embl.de/depod), protein kinases from UniProt (http://www.uniprot.org) and voltage-gated ion channels and solute carriers from the Human Gene Nomenclature Committee (http://www.genenames.org). Distributions were generated in R using knockdown data from the MCF10A and MCF10A-CDH1−/− cell line pair. The distribution of all 18,120 genes in the siRNA screen is represented by the blue lines (‘All’). The statistical significance of any differences between the distributions of all data (blue) and the functional group (red) was determined using the Kolmogorov-Smirnov test. KS p value = 0 equates to a p value of < 2.2 × 10−16. f Boxplot expanding the viability ratios of voltage-gated ion channels into functional subgroups. With the exception of the bestrophin, gap junction and leucine-rich subgroups, all groups were RSL, with a median viability ratio > 1. g–m Density distributions of cytoplasmic ribosomal proteins, spliceosome proteins, ubiquitin-specific peptidases, E2 ubiquitin-conjugating enzymes, HECT family ubiquitin E3 ligases, RING family ubiquitin E3 ligases and proteasome subunits. Lists were obtained from sources including ribosomal proteins—the ribosomal protein database (ribosome.med.miyazaki-u.ac.jp), ubiquitin-associated peptidases and E2 ubiquitin-conjugating enzymes—the Human Gene Nomenclature Committee (http://www.genenames.org), and E3 ubiquitin ligases from the National Heart Lung and Blood Institute (https://hpcwebapps.cit.nih.gov/ESBL/Database/E3-ligases/)
