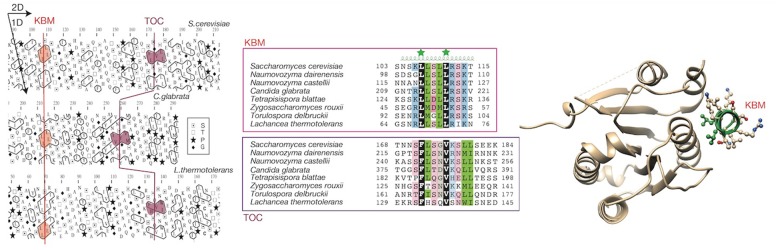
Fig. 2.
—The KBM–TOC region. Alignment of two conserved sequences defining the KBM and TOC motif. At left are shown the alignment of the 2D HCA plots of Sir4 from three species, as well as the corresponding 1D sequence alignments, extended to several other species. At right is shown the 3D structure of the recently solved KBM–KU complex (PDB 5Y59), with the two highly conserved leucine from KBM highlighted in green. On the HCA plots, the sequence is shown on a duplicated alpha helical net, on which the strong hydrophobic amino acids (V, I, L, M, F, Y, W) are contoured. These form clusters, which have been shown to mainly correspond to regular secondary structures (Gaboriaud et al. 1987; Callebaut et al. 1997). The way to read the sequence and the secondary structures are indicated with arrows. Special symbols are reported in the inset. In the alignment, positions conserved over the family are colored in green for hydrophobic amino acids (V, I, L, F, M, Y, W), in blue and pink for basic and acidic ones, in gray for loop forming residues (P, G, D, N, S). The UniProt identifiers of the sequences are reported in supplementary table S1, Supplementary Material online. No significant trace of KBM, moreover accompanied by a downstream TOC motif, could be found in the Kluyveromyces lactis and Eremothecium Sir4 sequences (both motifs presented in figure 3 of Chen et al. [2018] actually correspond to Asf2 sequences).