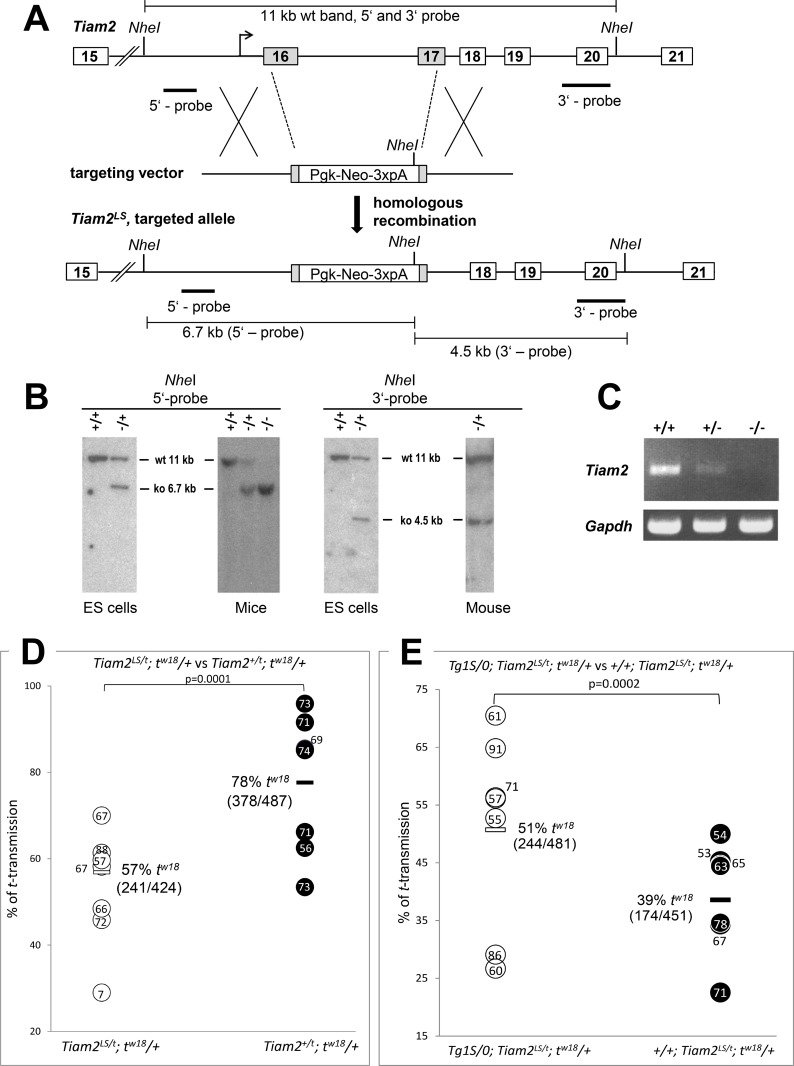
Fig 3. Targeted inactivation of the Tiam2 gene.
(A) Gene targeting strategy. Tiam2 was inactivated by introducing a neomycin selection cassette deleting exons 16 and 17. Introns are depicted as line, exons as boxes and targeted exons as shaded boxes. (B) Confirmation of correct homologous recombination. Southern blot analysis of NheI digested genomic DNA of ES-cells and mice with the right (3’) and left (5’) external probes. (C) Loss of Tiam2 transcription upon targeting of the Tiam2 locus as determined by RT-PCR expression analysis of both Tiam2l and Tiam2s in wild-type (+/+), heterozygous (+/-) and homozygous (-/-) mutant testes using a primer pair detecting both transcripts (see S6 Table). (D, E) t-haplotype transmission ratio of Tiam2LS/t males and Tiam2+/t control animals, without (D) or carrying in addition the transgene construct Tg1S (E). Horizontal bars denote average t-transmission ratio (%); the number of t carrying out of the total offspring is given in brackets below. Dots indicate the number of offspring sired by individual males. See also S4 and S5 Tables for total results. Abbr.: wt, wild-type; ko, knockout; +, wild-type allele;—, knock-out allele; 3xpA, polyadenylation signal.