Fig 3. Proteome analysis of the rcsB deletion effects.
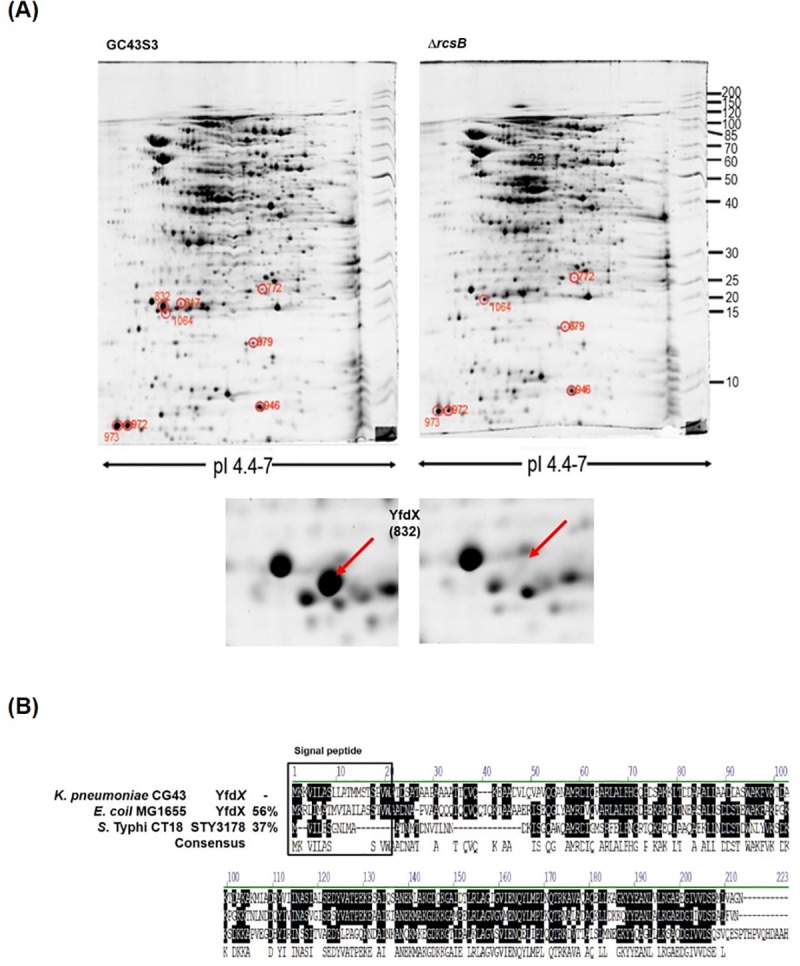
(A) Comparative proteome analysis of K. pneumoniae CG43S3 and CG43S3ΔrcsB. Representative SYPRO Ruby-stained gels derived from CG43S3 (WT) and ΔrcsB are shown. The exponential phase bacteria were incubated in LB broth at pH 4.4 for 1 h. proteome analysis was then performed. Spot 832, present only in CG43S3, was isolated and identified as YfdX through mass spectrometry. (B) Sequence comparison of YfdX of K. pneumoniae CG43S3, S. Typhi CT18, and E. coli MG1655. The predicted signal peptide (according to the SignalP 4.1 server) are marked.
