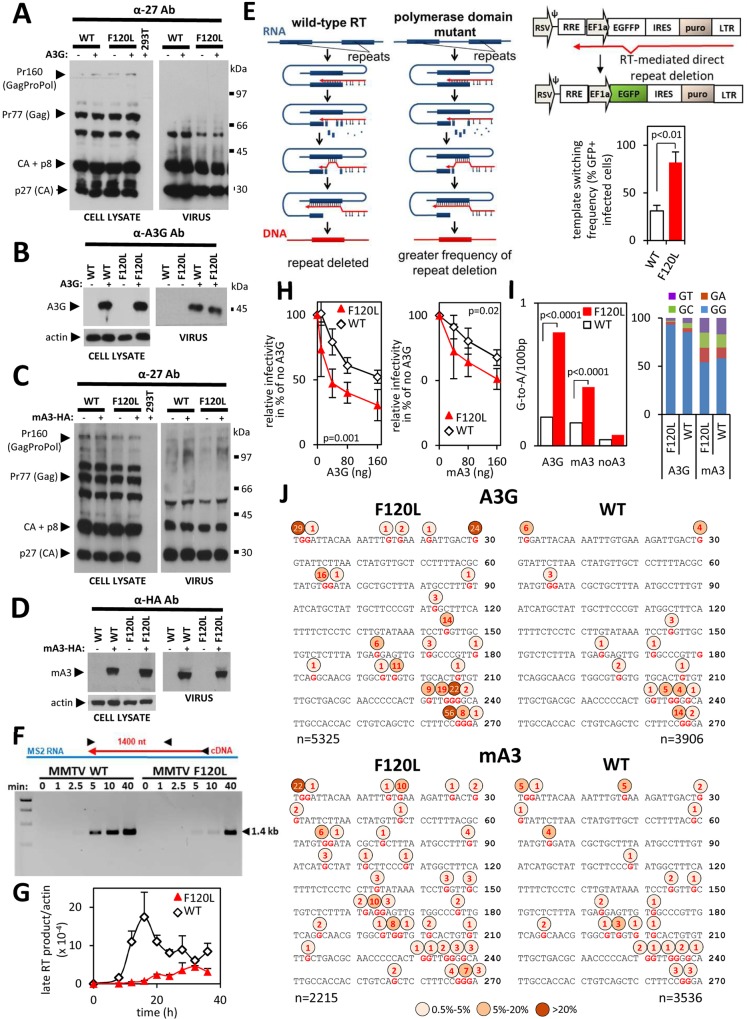
Fig 4. The F120L mutation, which does not affect A3s packaging, reduces the rate of DNA synthesis and sensitizes MMTV to inhibition by A3s.
(A—D) Detection of MMTV-specific proteins using anti-p27 (CA) antibody in cellular and viral lysates. For cellular lysates, 10 μg of total proteins obtained from MMTV WT or MMTV F120L virus-producing cells were used. Virus lysates were prepared from 100-fold concentrated virus preparations obtained from the supernatant of the MMTV WT or MMTV F120L virus-producing cells. The blots show unchanged expression levels and processing of capsid protein precursors following the introduction of the F120L mutation to the pol gene (A and C; left panel). Both, MMTV WT and MMTV F120L virions were also efficiently released from transfected cells (A and C; right panel). (B and D) Immunoblot detection of A3G and mA3 in the same lysates as shown in (A) and (C), respectively. An anti-actin antibody was used to show equivalent loading with cellular lysates. (A—D) One of four independent virus preparations, which were used for Fig 4H (right graph), I and J is shown. (E) A recombination assay. Direct repeats (thick blue line) are frequently deleted from retroviral genomes. Based on a dynamic copy choice model the deletion occurs when nascent minus DNA strand (red line) anneals to complementary RNA strand (blue line) situated upstream from the polymerizing reverse transcriptase (RT). The frequency of the deletion correlates with the length of the single-stranded (ss) DNA generated by RT. The longer ssDNA regions are generated with an RT exhibiting reduced activity of the DNA polymerase domain relative to the RNase H domain (polymerase domain mutants). Hindered DNA polymerization allows more efficient RNA degradation resulting in longer (-)ssDNA segments. The longer (-)ssDNA regions facilitate hydrogen bonding with the acceptor RNA template and in turn, enhance the recombination rate. The same mechanism applies to an egffp reporter gene carrying a direct repeat (f) in the central part of the egfp gene. Hydrogen bonding between nascent (-)ssDNA and viral RNA facilitates template switching and leads to deletion of the repeat. Reconstitution of the egfp gene results in the expression of functional EGFP that can be detected by UV microscopy and flow cytometry (right panel). Template switching frequency was determined for the MMTV WT RT and the MMTV F120L RT mutant. Differences between viruses were tested using Student’s two-tailed t-test (right panel) in GraphPad Prism. Results shown represent mean values with standard deviations from three experiments. A low MOI (<0.01) was used to ensure a single infection event per cell. A puromycin selection after infection ensures that only the infected cells are used for quantification of EGFP-expressing cells by flow cytometry. (F) The difference in the rate of DNA synthesis between the MMTV WT RT and MMTV F120L RT was analyzed using RTs liberated from virions, which were normalized to equal viral RNA levels (verified by EM, S3 Fig). The MS2 cDNA was synthesized and detected as described for Fig 3B. (marker: 1 kb DNA ladder, NEB). A representative Fig from three independent experiments is shown. (G) Accumulation of the late RT products in infected cells. DNA was extracted from 293T cells infected with MMTV WT or MMTV F120L virus preparations. DNA (100 ng) obtained at the indicated time points (0 h– 40 h) were subjected to a qPCR specific for the late MMTV RT products. Values were normalized to actin DNA levels. (H) mA3- and A3G-mediated inhibition of infectivity. Dose-response analysis of the inhibition of virus infectivity was performed as described in Fig 1D. Five independent virus preparations in five dose response infection experiments were performed for experiments with the mA3 (left). Four virus preparations were performed for the A3G tests (right). A two-way ANOVA in GraphPad Prism was used to analyze data and we found that the mutation significantly affects response to mA3 and A3G (I) Frequency of G-to-A editing found in the WPRE portion of the MMTV WT and MMTV F120L proviruses. The genomic DNA was extracted from cells infected with viruses produced in the absence (no A3G) or presence of A3G or mA3 (40 ng of A3 expression plasmid). A χ2 test performed in GraphPad Prism was used to analyze the differences in the frequency of deamination between viruses. (J) A G-to-A editing pattern detected in the plus DNA strands of the MMTV WT and MMTV F120L proviruses. Percent of mutated Gs at individual nt positions are depicted above the 270 bp consensus sequence.