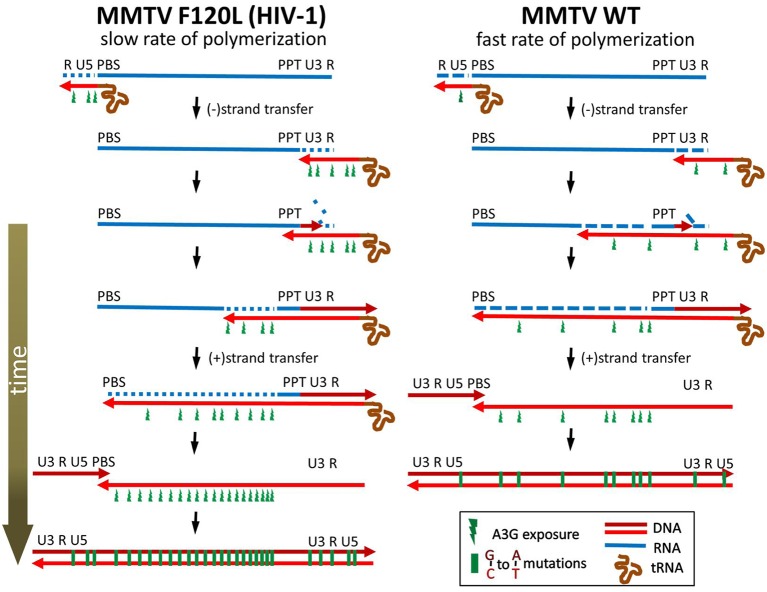
Fig 6. The proposed mechanism explaining lower sensitivity of MMTV to inhibition by A3s.
Reverse transcriptase (RT) produces the newly synthesized minus DNA strand (light red line) using viral RNA (blue line) as a template. The RNase H domain degrades the genomic RNA template behind the polymerizing DNA polymerase domain and exposes the minus DNA strand to A3s. Viruses containing RT exhibiting a fast rate of DNA polymerization (such as MMTV) narrow a window of opportunity for A3s to deaminate the single-stranded substrate as they complete synthesis of the double-stranded DNA in a shorter time compared to viruses carrying RT with a slow rate of polymerization (e.g. MMTV F120L or HIV-1). In addition, a reduction of the polymerase activity likely changes the balance between the polymerase and RNase H activity in favor of the later leading to the generation of more primary RNA cleavages per nucleotide addition and resulting in shorter RNA/DNA duplexes behind polymerizing RT. The viral RNA is blue; the dashed blue lines indicate RNase H cleavage; minus- and plus-strand DNA are light red and dark red, respectively. Single-stranded minus-sense DNA regions exposed to APOBEC3 are shown as green flashes.