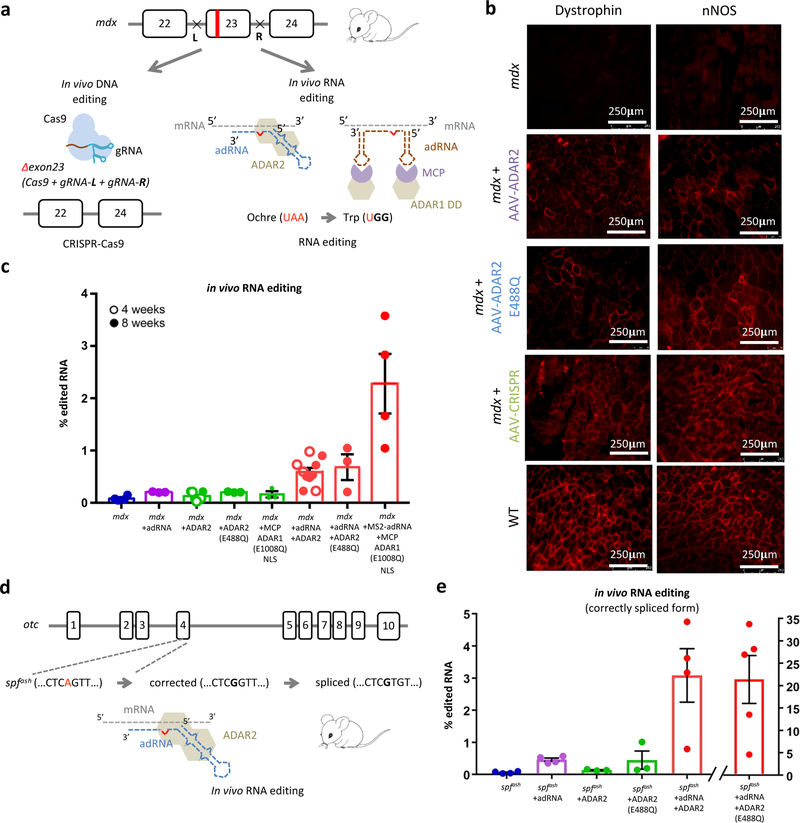
Figure 2: In vivo RNA editing in mouse models of human disease:
(a) Schematic of the DNA and RNA targeting approaches to restore dystrophin expression in the mdx mouse model of Duchenne Muscular Dystrophy: (i) a dual gRNA-CRISPR based approach leading to in frame excision of exon 23 and (ii) ADAR2 and MCP-ADAR1 based editing of the ochre codon. (b) Immunofluorescence staining for dystrophin in the TA muscle shows partial restoration of expression in treated samples (intra-muscular injections of AAV8-ADAR2, AAV8-ADAR2 (E488Q), and AAV8-CRISPR). Partial restoration of nNOS localization is also seen in treated samples (scale bar: 250μm). (c) In vivo TAA->TGG/TAG/TGA RNA editing efficiencies in corresponding treated adult mdx mice. Values represent mean +/− SEM (n=4, 3, 7, 3, 3, 10, 3, 4 independent TA muscles respectively). (d) Schematic of the OTC locus in the spfash mouse model of Ornithine Transcarbamylase deficiency which have a G->A point mutation at a donor splice site in the last nucleotide of exon 4, and approach for correction of mutant OTC mRNA via ADAR2 mediated RNA editing. (e) In vivo RNA correction efficiencies in the correctly spliced OTC mRNA in the livers of treated adult spfash mice (retro-orbital injections of AAV8-ADAR2 and AAV8-ADAR2 (E488Q)). Values represent mean +/− SEM (n=4, 4, 3, 3, 4, 5 independent animals respectively).