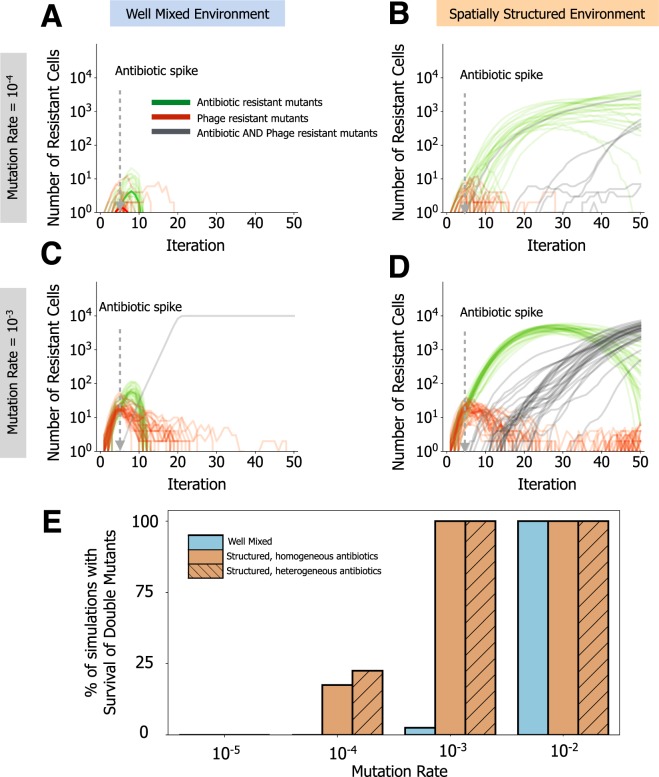
Figure 3.
Spatial structure promotes the emergence of multi-resistant bacteria. (A–D) Simulations of a single bacterial species, initially sensitive to antibiotics and phage, evolving in the presence of both stressors. Lines show 30 replicate simulations with emerging resistant lineages (to one or both selective pressures). Single mutants resistant to phage are shown in red, whilst single mutants resistant to bacteria are shown in green. Double mutant lineages resistant to antibiotics and phage are shown in grey. Antibiotics are applied at the time point indicated by the arrow, whilst phages (10 particles) are inoculated at iteration 0 along with bacteria. In (A and B) resistant mutants emerge at a rate of 10−4 per cell per iteration (C and D) resistant mutants emerge at a rate of 10−3 per cell per iteration. (A and C) show dynamics from well-mixed environments. (B and D) show dynamics from spatially structured environments. (E) Percentage of simulations (out of 30) where lineages resistant both to antibiotics and phage have emerged, in either well mixed or spatially structured environments (with homogeneous or heterogeneous antibiotic exposure), for all the mutation rates tested (x-axis). The complete set of parameters for these simulations is show in Supplementary Data 1.