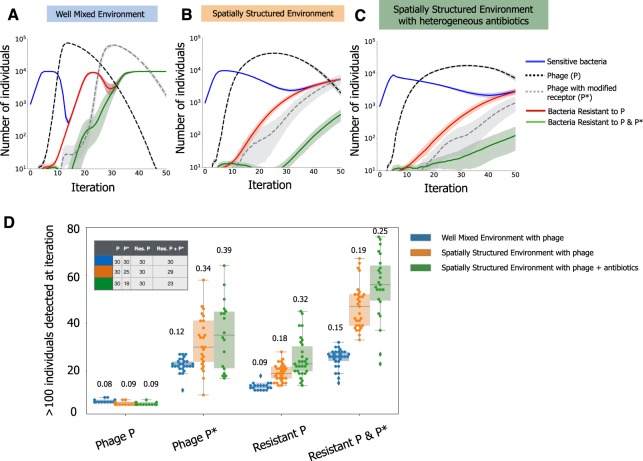
Figure 4.
Co-evolutionary arms-race between bacteria and phage across different environments. In these simulations, bacteria can only become resistant to phage by mutating its receptor. Accordingly, phage can re-infect bacteria with a mutated receptor phenotype by mutating its own receptor. Sensitive bacteria (full blue line) are co-inoculated with virulent phages (dashed black line). Mutations conferring resistance to a random phage receptor phenotype occur at a rate of 10−3 per generation, and can give rise to bacterial mutants resistant to one (full red line) or two (full green line) phage receptor phenotypes. Mutations randomly changing the attachment morphology of phage P occur at 10−4, and can give rise to a phage with a different attachment phenotype (P*, dashed grey line). Opaque bands indicate the 95% confidence interval. (A) Simulations in well-mixed environments. (B) Simulations in spatially structured environments. (C) Simulations in spatially structured environments supplemented with antibiotics, that are applied heterogeneously. Here antibiotic resistant mutants emerge with probability 10−3. (D) The distribution of the iteration, in replicate simulations, at which given phage or bacterial genotypes were detected in at least 100 individuals, for the different treatment regimes in (A–C). Each point indicates a different simulation. The number of replicate simulations (out of 30) where the genotypes were not detected until the final time point (80 iterations) are not included in the distributions. Instead, the inset table shows the number of simulations where each phenotype has emerged in the different conditions (e.g., counter-resistance phage P* emerged only in 18 out of 30 replicate simulations performed in spatially structured environments complemented with heterogeneous antibiotics). Numbers above each distribution indicates the respective coefficient of variance, calculated based only on the simulations where the respective genotype was detected. The complete set of parameters used is shown in Supplementary Data 1.