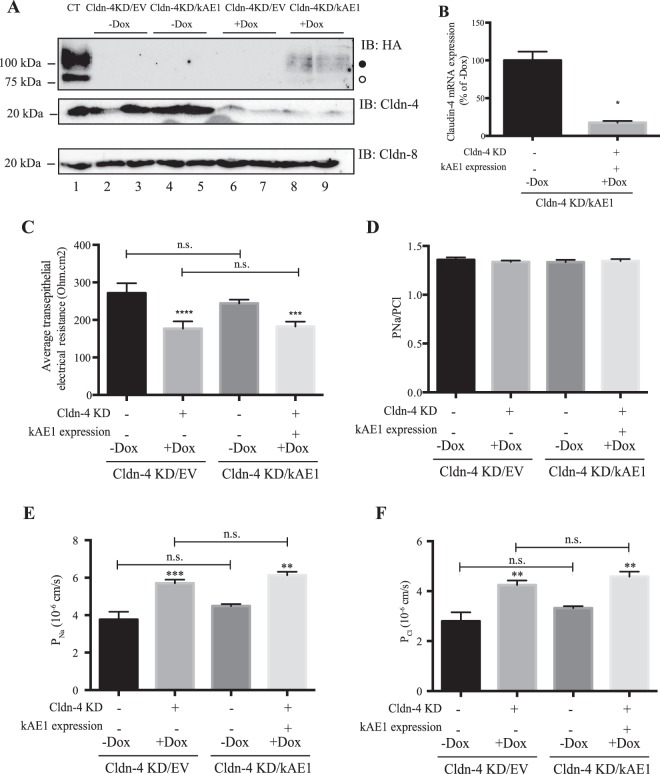
Figure 6.
Claudin-4 knockdown reduces TEER and increases absolute permeability to both sodium and chloride. (A) mIMCD3 cells expressing kAE1, Cldn-4/EV or Cldn-4/kAE1 in an inducible manner were either incubated in presence or absence of doxycycline for 48 hours, prior to lysis and analysis of protein abundance by immunoblot. Mouse anti-HA antibody was used to detect kAE1 protein (upper panel), rabbit anti-claudin-4 (middle panel) or rabbit anti-claudin-8 (bottom panel) antibodies were used respectively to detect claudin-4 and claudin-8. The dark circle corresponds to kAE1 carrying complex oligosaccharide, and the white circle indicates kAE1 carrying high mannose oligosaccharide. Twenty micrograms of proteins were loaded in every lane. (B) Quantitative PCR analysis of claudin-4 mRNA abundance in Cldn-4 KD/kAE1 incubated in control conditions or with Doxycycline. The results are expressed as a percentage of “Cldn-4 KD/kAE1 – Dox” conditions and are normalized to the expression of actin. Error bars correspond to means ± SEM, n = 3. *P < 0.05 versus “Cldn-4 KD/kAE1 - Dox” condition using un-paired t-test. (C) Ussing chambers data showing an equivalent decrease in TEER in both Cldn-4 KD/EV and Cldn-4/kAE1 cells incubated with doxycycline. Error bars correspond to means ± SEM, n = 4. ***P < 0.001 versus Cldn-4 KD/kAE1 cells without doxycycline, ****P < 0.0001 versus Cldn-4 KD/EV cells without doxycycline. n.s. indicates non significant difference (One-way ANOVA). Although PNa / PCl ratio was unchanged, (D), absolute permeability to both sodium (PNa) (E) or chloride (PCl) (F) were increased upon Doxycycline incubation of Cldn-4/EV and Cldn-4/kAE1 cells. Error bars correspond to means ± SEM, n = 4, ***P < 0.001 versus Cldn-4 KD/EV cells without doxycycline, **P < 0.01 versus the same cell line without doxycycline.