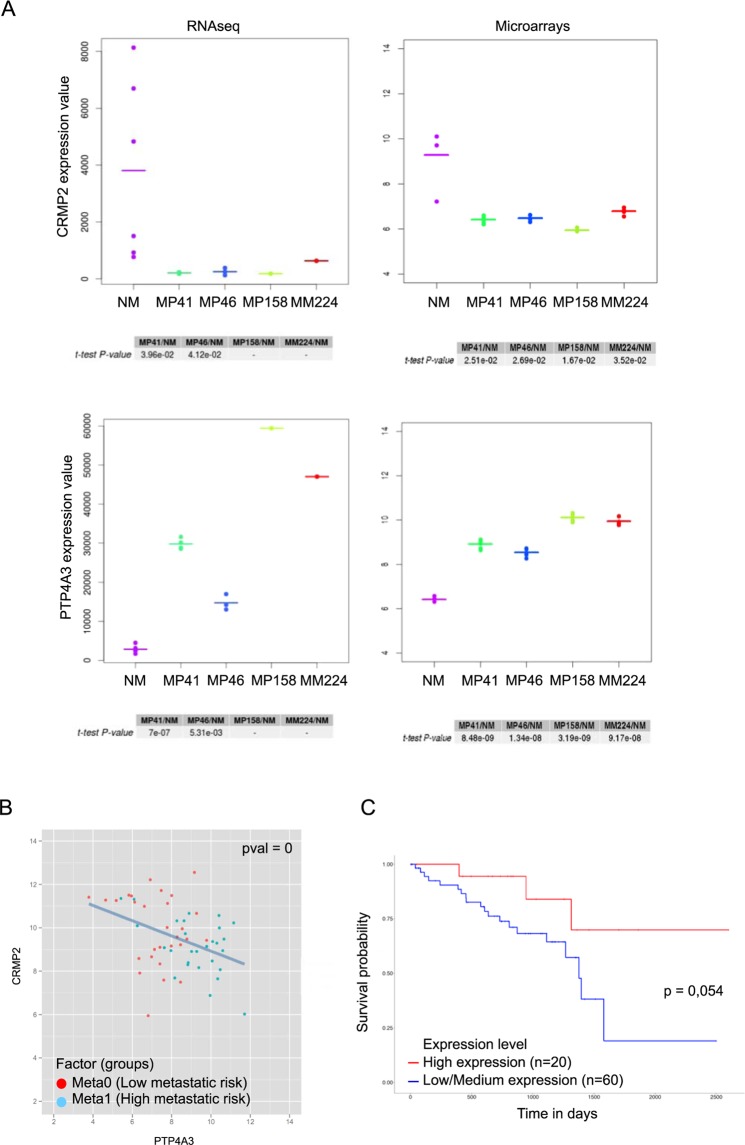
Figure 6.
Expression level of PTP4A3 and CRMP2 in normal melanocytes, uveal melanoma cell lines, tumors and survival probability in patients. (A) A Transcriptome analysis was conducted on replicates of normal melanocytes (NM), and PDX of primary tumors such as MP41, MP46, MP158, and a PDX of a metastasis MM224. Normal melanocytes, and uveal melanoma tumor cells from cell sorting of dissociated PDX were analyzed on Affymetrix Human Transcriptome Array v2.0 and on Illumina to performed gene expression analysis48. Signal from microarray data were normalized according Genosplice’s pipeline based on a RMA normalization step as described previously49. RNAseq signals are Deseq. 2-Normalized counts processed by Genosplice’s pipeline too50. A paired t-test was applied between UM and normal melanocytes. In our comparisons, CRMP2 is down regulated in UM vs NM, as observed in RNASeq and in microarray datasets. PTP4A3 is upregulated in UM vs normal melanocytes. No p-value are calculated for MP158 and MM224 in RNAseq dataset because a unique sample was sequenced contrary to first experiments done on microarrays. MP41 and MP46 were described previously23,24. MP158 and MM224 are GNAQ mutated and BAP-1 mutated. (B) PTP4A3 and CRMP2 expression were determined by microarrays analysis. Specimens were analyzed on GeneChip Human Genome U133 Plus 2.0 microarrays (Affymetrix) as described previously7. PTP4A3 and CRMP2 expression are inversely correlated in tumors (Spearman coefficient of −0,4). Spearman instead of Pearson correlation, because the Spearman correlation is less sensitive than the Pearson correlation to strong outliers. 63 tumors were analysed, each one divided in two groups: the red dots correspond to meta0 tumors (low metastatic risk) and blue dots to meta1 tumors (high metastatic risk)7. (C) Effect of CRMP2 expression on UM survival. Kaplan-Meier analysis of CRMP2 expression in 80 uveal melanoma tumors from the TCGA database (http://ualcan.path.uab.edu/index.html).