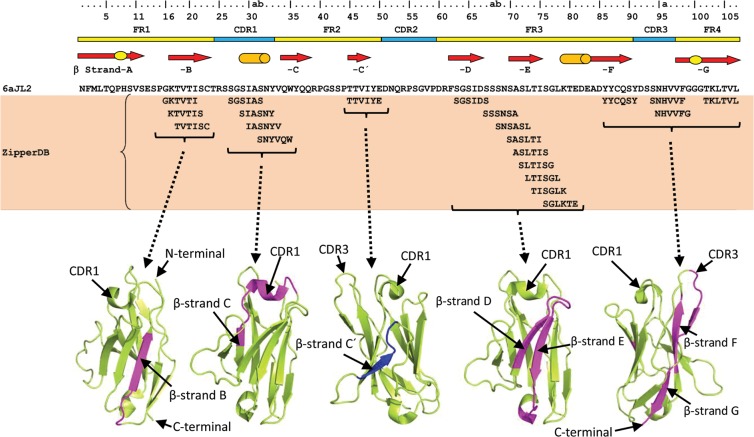
Figure 1.
Segments of 6aJL2 protein predicted to be fibrillogenic by the computational tool ZipperDB26,29 (https://services.mbi.ucla.edu/zipperdb/). The hexamers shown are those with a Rosetta energy ≤−23 kcal/mol, which are predicted to form fibrils27. The location of the clusters of the amyloidogenic hexamers is shown highlighted in magenta in the three-dimensional structure of 6aJL2 protein (bottom). The regions of 6aJL2 protein with β-strands or helix conformation in the native state are indicated by arrows and cylinders, respectively (Top). The oval figures in the first and last arrows represent, respectively, the sheet-switch motif characterizing the structure of the N-terminal segment, and the β-bulge centred at Gly100 in the β-strand G. The residue numbering and the location of the CDR/FR regions are according to Chothia and Lesk69. The graphical representations of 6aJL2 structure were prepared with PyMOL70, based on the structure contained in the PDB 2W0K.