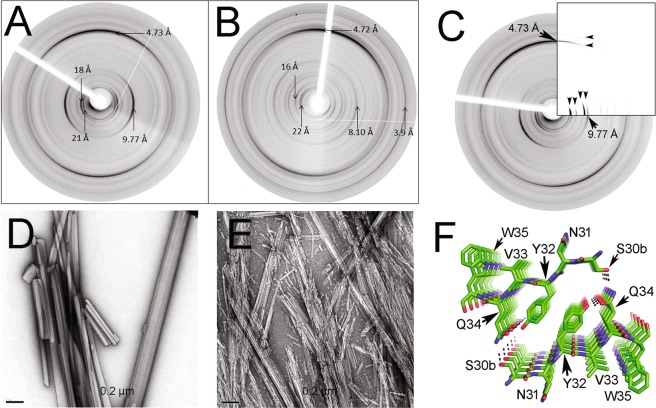
Figure 5.
X-ray fibre diffraction (XRFD) patterns from partially aligned fibrils formed by synthetic peptides (A) Ser30b-Trp35 and (B) Ile30-Gln37 incubated in MilliQ-quality water plus Na azide 0.05%, as described in Methods. (D,E) show transmission electron micrographs of the aggregates whose XRFD pattern are shown in panels A and B, respectively. The images were obtained with a HITACHI-7100 transmission electron microscope (C). Comparison of the experimental XRFD pattern of the aggregates of the peptide Ser30b-Trp35, as shown in (A), and the simulated pattern (inserted quadrant) calculated based on the theoretical model shown in (F). Black arrows are shown to highlight positions of matching major diffraction signal positions on the equator and meridian. (F) Structural model of the steric zipper of peptide Ser30b-Trp35, generated by the computational tool ZipperDB26. The amino acid side chains are shown in stick representation and named with the one-letter code. The dotted lines represent intra- and interpeptide H-bonds stabilizing the crystal lattice. Figure prepared with PyMOL70.