Figure 6.
The Structure and Mechanism of the Dehydrogenase McbC
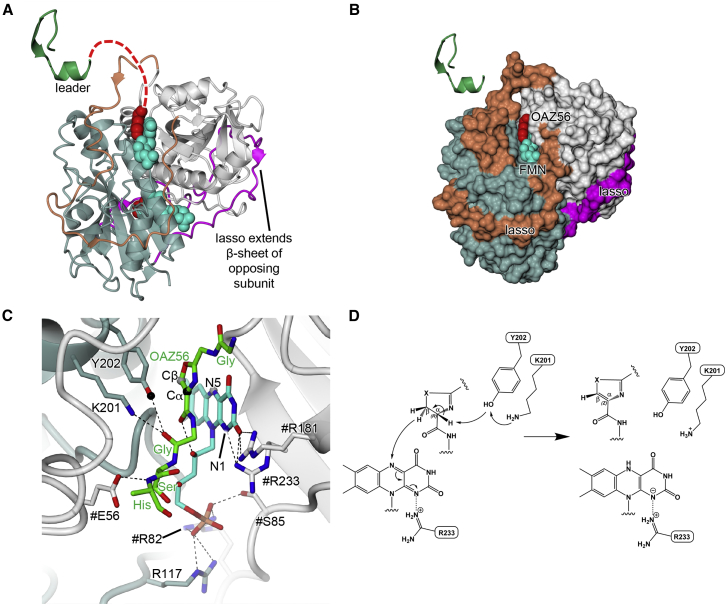
(A and B) show the isolated McbC dimer (in standard view) in cartoon (A) and molecular surface (B) representations, respectively. McbC and its lasso are colored slate blue and magenta, respectively, while #McbC and its lasso are colored gray and orange, respectively. Also shown is the relative position of the leader peptide (green) and, as van der Waals spheres, the oxazole (OAZ56; red) bound alongside the FMN cofactor (cyan). A putative path for the substrate peptide extending from the end of the leader and going toward the McbC active site is traced by the red dashed line in (A).
(C) Close up of the active site region showing side chains (in stick representation) that interact with the FMN and bound product. The black spheres indicate the atoms involved in the deprotonation step, and the gray spheres indicate the atoms involved in the hydride transfer.
(D) Proposed general mechanism for McbC: an activated Tyr202 abstracts a proton from the α carbon of an azoline substrate, which results in E2 elimination of the antiproton from the β carbon and hydride transfer to FMN. Instead of a proton being provided to FMN by a general base to yield FMNH2, we propose that the negative charge on N1 of FMN is stabilized by a salt bridge with Arg233.