Abstract
MicroRNA-212 (miR-212) is dysregulated in numerous tissues and cancer types and serves a role in the progression of human cancer. However, the function and mechanism of miR-212 in the development of pancreatic ductal adenocarcinoma (PDAC) remain unknown, particularly in a hypoxic microenvironment. In the present study, miR-212 expression was observed to be significantly upregulated in PDAC tissues compared with normal tissues. Clinical data analysis indicated that miR-212 was positively associated with a large tumor size, Tumor-Node-Metastasis stage, lymph node metastasis and vessel invasion, and influenced the overall survival time. Notably, there was a positive association between the expression of hypoxia-inducible factor-1α (HIF-1α) and miR-212 in vivo and in vitro in hypoxic conditions. Mechanistically, HIF-1α bound directly to a hypoxia response element in the miR-212 promoter region and activated miR-212 expression in PDAC cells. Collectively, these results demonstrated that HIF-1α positively regulated miR-212 expression and resulted in PDAC progression.
Keywords: pancreatic cancer, microRNA-212, hypoxia-inducible factor-1α, progression
Introduction
Pancreatic ductal adenocarcinoma (PDAC) is the fourth leading cause of cancer-associated mortality in the USA and likely to rise to the second by the year 2030 (1). The 5-year overall survival rate is only 8.0% and the median survival time of patients is <6 months (2). Despite rapid improvements in PDAC diagnosis and therapy, the overall survival time of patients with pancreatic cancer has not changed substantially (3). Patients with PDAC mostly succumb due to recurrence and metastasis to distant organs (4). Furthermore, there remains a lack of underlying biomarkers and therapeutic targets for PDAC diagnosis and therapy. Therefore, it is necessary to identify novel biomarkers and therapeutic targets in order to improve the efficacy of treatment for patients with PDAC.
MicroRNAs (miRNAs/miRs) are small noncoding RNAs, usually 18–25 nucleotides in length, which regulate protein translation by binding to the 3′-untranslated region of their target mRNAs (5). miRNAs serve roles in numerous physiological and pathological processes including apoptosis, the cell cycle, cellular growth, proliferation, differentiation, metabolism and aging (6). Accumulating evidence reveals that miRNA dysfunction influences the metastasis, chemo-radioresistance and overall survival time of patients with cancer, including patients with PDAC (7–10). Previous reports have revealed miRNA involvement in PDAC progression, including roles of miR-216b (11), miR-10a-5p (10), miR-374b-5p (12), miR-1271 (13), miR-7-5p (14), miR-4295 (15) and miR-185 (16). However, the molecular mechanisms underlying the involvement of miRNAs in PDAC progression remain unclear.
miR-212, a cancer-associated miRNA located on chromosome 17p13.3, participates in the progression of different types of human cancer (17). It has been demonstrated that miR-212, as an oncogene, is upregulated in breast cancer (18) and pancreatic cancer (19); and as a suppressor gene, is downregulated in hepatocellular carcinoma (20), prostate cancer (21), non-small cell lung cancer (22) and nasopharyngeal carcinoma (23). Furthermore, aforementioned studies revealed that dysregulated miR-212 has an influence on the progression of a number of solid malignancies. However, to the best of our knowledge, the function and regulatory mechanism of miR-212 in PDAC has not yet been examined.
The present study aimed to investigate the function and expression mechanism of miR-212 in PDAC. The results revealed that miR-212 was overexpressed in PDAC specimens and associated with certain clinicopathological features, particularly the overall survival time of patients. miR-212 overexpression in PDAC was revealed to be mediated by hypoxia-inducible factor-1α (HIF-1α), which directly bound to the miR-212 promoter and upregulated its activity. The results of the present study indicate that miR-212 is a novel direct target of HIF-1α and may function as a novel biomarker for the diagnosis and therapy of PDAC.
Materials and methods
Cell culture and hypoxic treatment
Human PDAC cell lines MiaPaca2 and AsPc1, and the 293 cell line, were obtained from the Cell Bank of Type Culture Collection of Chinese Academy of Sciences (Shanghai, China). These cells were cultured at 37°C in a humidified atmosphere of 5% CO2 using Dulbecco's modified Eagle's medium with 10% fetal bovine serum (both Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA). For hypoxic treatment, cells were placed in a modulator incubator chamber (Thermo Fisher Scientific, Inc.) with 93.5% N2, 5% CO2 and 1.5% O2 for 6 and 12 h. Oxygen at 21% was used during normoxia condition.
With an approval from the Ethics Committee of the Tianjin Cancer Institute and Hospital (Tianjin, China), PDAC and adjacent normal pancreatic tissue specimens were obtained from patients (age, 51–75 years; male, 12; female, 29) who underwent surgical resection subsequent to the histopathological diagnosis of PDAC (n=41). Patients were included if they were diagnosed with PDAC via pathological diagnosis following radical resection and if they were willing to participate. Patients were excluded if they did not undergo radical resection. Written informed consent was obtained from individuals or their guardians. Samples were collected between January and December 2015 at Tianjin Cancer Institute and Hospital with a follow-up period of 2 years to analyze over survival.
miR-212 structure search
miRbase (www.mirbase.org) was searched using ‘miR-212’ and MI0000288 was selected to identify precursor and mature miR-212 structures. The GC box and the TATA box located at the transcription initiation point of miR-212 were identified using the University of California Santa Cruz genome browser (genome.ucsc.edu) by selecting genomes and searching ‘miR-212’ and selecting ‘MIR212 (ENST00000586026.1)’ for further studies. Five hypoxia response elements (HREs) were identified (5′-A/GCGTG-3′) to potentially to bind HIF-1α at the miR-212 promoter.
Small interfering RNA (siRNA), plasmid constructs and transient transfection
siRNAs against HIF-1α (siHIF-1α) were designed and synthesized by Shanghai GenePharma Co., Ltd. (Shanghai, China), including the following duplexes: siHIF-1α-1, forward, 5′-CCAGCAGACUCAAAUACAATT-3′ and reverse, 5′-UUGUAUUUGAGUCUGCUGGTT-3′; siHIF-1α-2, forward, 5′-GCCUCUUUGACAAACUUAATT-3′ and reverse, 5′-UAAGUUUGUCAAAGAGGCTT-3′; siHIF-1α-3, forward, 5′-CAGGCCACAUUCACGUAUATT-3′ and reverse, 5′-UAUACGUGAAUGUGGCCUGTT-3′; and siNC, forward, 5′-UUCUCCGAACGUGUCACGUTT-3′ and reverse, 5′-ACGUGACACGUUCGGAGAATT-3′. Following the examination transfection efficacy, siHIF-1α-3 was selected for subsequent experiments. The human HIF-1α cDNA was cloned into the pcDNA3.1 plasmid expression vector (Thermo Fisher Scientific, Inc.) for overexpression experiments and the empty vector was used as a control. siRNAs (50 nM) and plasmids (2 µg) were prepared and transfected into the PDAC cell lines using Lipofectamine® 2000 (Thermo Fisher Scientific, Inc.) (18,19). At 48 h of transfection, mRNA and protein levels were tested by reverse transcription-quantitative polymerase chain reaction (RT-qPCR) and western blotting.
RT-qPCR
Total RNA was isolated from the transfected cells using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.) and used for first-strand cDNA synthesis (42°C for 20 min; 95°C for 5 min; and 5°C for 5 min) using the First-Strand Synthesis System for RT-PCR (Takara Biotechnology Co., Ltd., Dalian, China). Each sample was processed in triplicate using a SYBR-Green PCR kit (cat no RR820A; Takara Biotechnology Co., Ltd.) with the following thermocycling conditions: 95°C for 30 sec followed by 40 cycles of 95°C for 5 sec and 60°C for 30 sec. Relative mRNA expression levels were quantified using the 2−ΔΔCq method (24). U6 RNA and β-actin were used as reference genes. The following primers were obtained from Shanghai GenePharma Co., Ltd. (Shanghai, China): has-miR-212 sense, 5′-ACACTCCAGCTGGGTAACAGTCTCCAGTC-3′; HIF-1α sense, 5′-AGAAACCACCTATGACCTGC-3′ and antisense, 5′-GTCGTGCTGAATAATACCACTC-3′; U6 sense, 5′-GCTTCGGCAGCACATATACTAAAAT-3′ and antisense, 5′-CGCTTCACGAATTTGCGTGTCAT-3′; β-actin sense, 5′-CTGGAACGGTGAAGGTGACA-3′ and antisense, 5′-AAGGGACTTCCTGTAACAATGCA-3′.
Western blot analysis
Whole-cell extracts were prepared by lysing cells with radioimmunoprecipitation assay lysis buffer in a proteinase inhibitor cocktail (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany). Protein concentrations were quantified using a bicinchoninic acid protein assay kit (Pierce; Thermo Fisher Scientific, Inc.). Protein lysates (20 µg/lane) were separated on 10% SDS-PAGE gels and transferred to polyvinylidene fluoride membrane. Membranes were blocked with PBS plus 0.1% Tween-20 with 5% non-fat milk at room temperature for 2 h. Target proteins were detected by probing with antibodies against HIF-1α (1:1,000; cat. no. ab16066; Abcam, Cambridge, UK) and β-actin (1:1,000; cat. no. ab8226; Abcam) overnight at 4°C. Following, membranes were incubated with anti-mouse IgG second antibody (1:1,000; cat. no. ab190475; Abcam) at room temperature for 2 h. Specific proteins were visualized using an enhanced chemiluminescence detection reagent (cat. no. #32106; Pierce; Thermo Fisher Scientific, Inc.).
Chromatin immunoprecipitation (ChIP) assay
A chromatin immunoprecipitation assay was performed using a commercial kit (cat no. #17-295; Upstate Biotechnology, Inc., Lake Placid, NY, USA) according to the manufacturer's protocol. MiaPaca2 and 293 cells were treated under hypoxia conditions (93.5% N2, 5% CO2 and 1.5% O2) in the modulator incubator chamber at 37°C for 6 and 12 h. Oxygen at 21% was used during normoxia. ChIP assays were performed to confirm HREs that bind to HIF-1α. The following groups were analyzed: Negative control (NC), positive control (PC), normoxia (N) and hypoxia (H).
Dual-luciferase reporter assay
Genomic DNA fragments of the miR-212 gene, spanning from +1 to −2,000 relative to the transcription initiation sites were generated by PCR and inserted into pGL3-Basic vectors (pGL3-miR-212; cat. no. E1751; Promega Corporation, Madison, WI, USA). A mutation (mut) HRE of miR-212 was created for dual-luciferase reporter assays in addition to wild type fragments. All constructs were sequenced to confirm their identity. Plasmids (2 µg) were transfected into MiaPaca2 and 293 cells using Lipofectamine® 2000 (Thermo Fisher Scientific, Inc.). At 48 h of transfection, cells were prepared for Dual-Luciferase Reporter assays. Luciferase activity was measured using the Dual-Luciferase Reporter assay system (Promega Corporation) as previously described (25). Renilla activity was used for normalization.
Statistical analysis
Differences between groups were compared using a Student's t-test or one-way analysis of variance followed by a least significant difference post hoc test. Categorical data were analyzed using either Fisher's exact test or the χ2 test, as appropriate. Each experiment was conducted independently at least three times, and the values are presented as the mean ± standard error of the mean unless otherwise stated. Statistical analyses were performed using SPSS software (version 21.0; IBM Corp., Armonk, NY, USA). P<0.05 was considered to indicate a statistically significant difference.
Results
miR-212 and HIF-1α mRNA are overexpressed in PDAC and associated with the clinicopathological features and prognosis of patients with PDAC
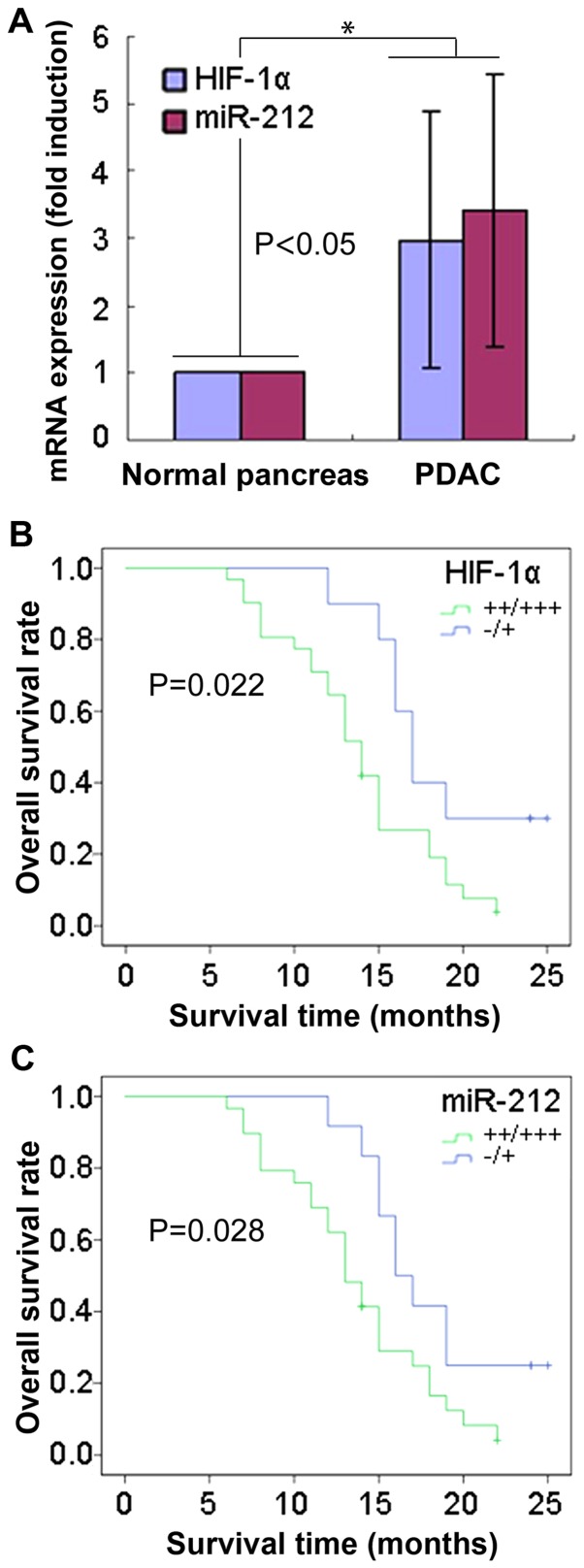
The expression level of miR-212 and HIF-1α mRNA was detected by RT-qPCR using paired specimens from patients with PDAC. The data indicated that miR-212 and HIF-1α mRNA expression levels were significantly upregulated in PDAC samples compared with adjacent normal pancreatic tissue samples (P<0.05; Fig. 1A). There was a positive association identified between miR-212 and HIF-1α at the mRNA level. Subsequently, miR-212 expression level in PDAC samples was assessed by RT-qPCR and it was revealed that there was a significant association between miR-212 expression level and tumor size, lymph node metastasis and vessel invasion among patients with PDAC (all P<0.05; Table I). miR-212 was quartered according to the range of miR-212 expression, <25% was considered as negative expression (−), 26–50% was low expression (+), 51–75% was termed medium expression (++) and >76% identified high expression (+++). Patients with PDAC with a high expression level of miR-212 and HIF-1α mRNA had a significantly worse overall survival time compared with patients with a low expression level (P=0.022 and P=0.028, respectively; Fig. 1B and C), suggesting that miR-212 and HIF-1α may serve a role in the survival of patients with PDAC. The results revealed that miR-212 and HIF-1α are overexpressed in PDAC samples and their expression was associated with clinicopathological features in PDAC, particularly the overall survival time of patients.
Figure 1.
miR-212 and HIF-1α mRNA expression levels in samples from patients with PDAC and the association with prognosis. (A) Expression analysis of miR-212 and HIF-1α mRNA levels in PDAC samples and adjacent normal pancreatic samples determined by a reverse transcription-quantitative polymerase chain reaction. (B) Association between HIF-1α expression levels and the overall survival time of patients with PDAC. (C) Association between miR-212 expression levels and the overall survival of patients with PDAC. P<0.05 was determined using the log-rank test. *P<0.05. miR, microRNA; HIF-1α, hypoxia-inducible factor-1α; PDAC, pancreatic ductal adenocarcinoma; (−), negative expression (<25%); (+), low expression (26–50%); (++) medium expression (51–75%); (+++) high expression (>76%).
Table I.
Association between miR-212 expression and clinicopathological features in patients with pancreatic ductal adenocarcinoma.
| miR-212 | ||||
|---|---|---|---|---|
| Variables | −/+ | ++/+++ | χ2 test | P-value |
| Tumor size (cm) | 10.041 | 0.004 | ||
| T1 | 6 | 2 | ||
| T2 | 6 | 27 | ||
| Histological grade | 0.277 | 0.702 | ||
| G1/G2 | 10 | 22 | ||
| G3 | 2 | 7 | ||
| pTNM stage | 2.305 | 0.165 | ||
| I/II | 10 | 17 | ||
| III/IV | 2 | 12 | ||
| Lymph node metastasis | ||||
| N0 | 7 | 6 | 5.555 | 0.029 |
| N1 | 5 | 23 | ||
| Vessel invasion | ||||
| Negative | 6 | 5 | 4.640 | 0.041 |
| Positive | 6 | 24 | ||
| HIF-1α | 16.443 | <0.001 | ||
| −/+ | 8 | 2 | ||
| ++/+++ | 4 | 27 | ||
P-values were calculated using a χ2 test. miR, microRNA; HIF-1α, hypoxia-inducible factor-1α; pTNM, pathological Tumor-Node-Metastasis; (−), negative expression (<25%); (+), low expression (26–50%); (++) medium expression (51–75%); (+++) high expression (>76%)
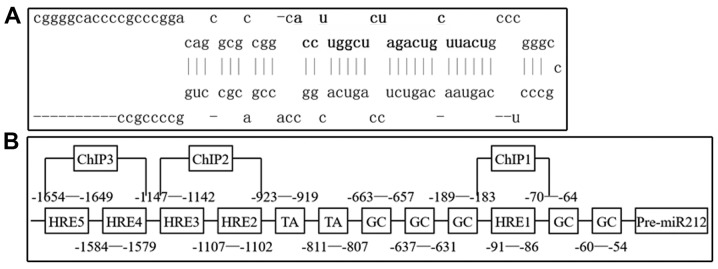
Structure of miR-212
The present study used the miRBase database to identify the precursor and mature miR-212 structures (Fig. 2A). The GC box and the TATA box located at the transcription initiation point of miR-212 were identified using the University of California Santa Cruz genome browser (Fig. 2B) and a total of five HREs were identified (5′-A/GCGTG-3′) that were potentially to bind with HIF-1α at the miR-212 promoter (Fig. 2B). Therefore, HIF-1α may bind to the HRE of miR-212 to initialize the transcription of miR-212.
Figure 2.
Structure of miR-212. (A) Using the miRBase database, the miR-212 precursor (upper) and mature (lower) structures were identified. (B) The GC box and the TATA box were located at the transcription initiation point of miR-212 and there were 5 HERs (5′-A/GCGTG-3′) identified. miR, microRNA; HER, hypoxia response element; ChIP, chromatin immunoprecipitation assay.
miR-212 and HIF-1α mRNA levels were upregulated under in vitro hypoxic conditions
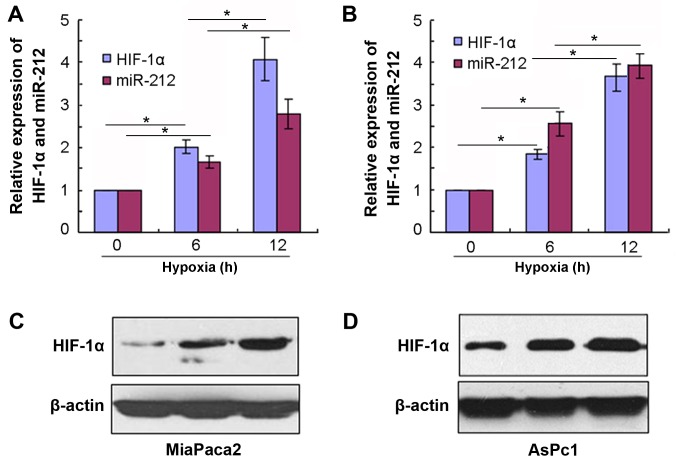
To further confirm the positive association between miR-212 and HIF-1α mRNA expression, the pancreatic cancer cell lines (MiaPaca2 and AsPc1) were used for in vitro experiments in a hypoxic microenvironment. MiaPaca2 and AsPc1 cell lines were maintained in hypoxic conditions for different durations (6 and 12 h) and RT-qPCR was used to assess the mRNA expression of miR-212 and HIF-1α. The results indicated that the expression levels of miR-212 and HIF-1α mRNA were significantly upregulated following hypoxia stimulation at 6 h compared with 0 h and at 12 h compared with 6 h (P<0.05; Fig. 3A and B). Additionally, HIF-1α protein expression levels were markedly increased in hypoxia conditions at different time points in MiaPaca2 and AsPc1 cells (Fig. 3C and D). Furthermore, it was revealed that miR-212 and HIF-1α mRNA expression levels were positively associated in MiaPaca2 and AsPc1 cell lines in hypoxic milieu 12 h.
Figure 3.
miR-212 and HIF-1α mRNA expression levels are upregulated in in vitro under hypoxic conditions. (A) miR-212 and HIF-1α mRNA expression levels were measured using RT-qPCR following hypoxic stimulation in MiaPaca2 cells. (B) miR-212 and HIF-1α mRNA expression levels were measured using RT-qPCR following hypoxic stimulation in AsPc1 cells. HIF-1α protein expression levels were measured by western blotting following hypoxic stimulation in (C) MiaPaca2 and (D) AsPc1 cell lines. *P<0.05. miR, microRNA; HIF-1α, hypoxia-inducible factor-1α; RT-qPCR, reverse transcription-quantitative polymerase chain reaction.
HIF-1α influences the expression of miR-212
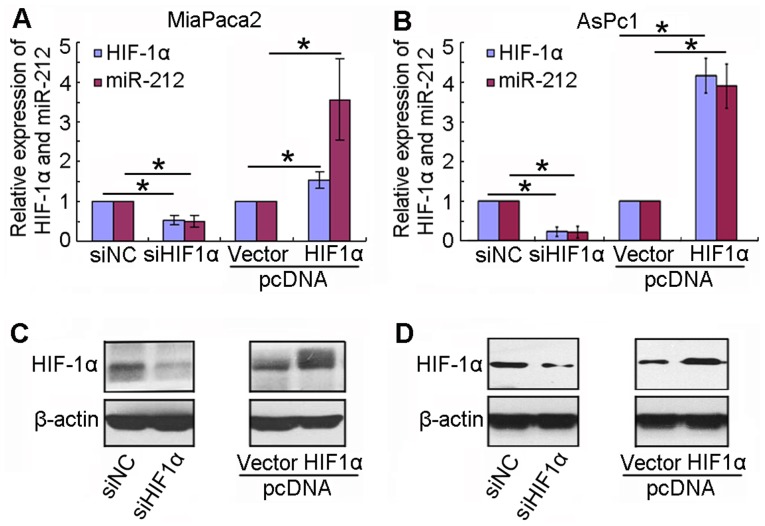
In order to confirm the influence of HIF-1α on miR-212 expression, siHIF-1α and HIF-1α plasmid were transfected into MiaPaca2 and AsPc1 cell lines. The results indicated that miR-212 expression was significantly downregulated in MiaPaca2 and AsPc1 cell lines transfected with siHIF-1α compared with control cells (P<0.05; Fig. 4A and B). By contrast, miR-212 expression was significantly upregulated in MiaPaca2 and AsPc1 cell lines transfected with the HIF-1α overexpression plasmid compared with control cells (P<0.05; Fig. 4A and B). Fig. 4C and D presents the protein expression levels of HIF-1α subsequent to transfection with siHIF-1α and HIF-1α plasmid in MiaPaca2 and AsPc1 cell lines, respectively. The data implied that HIF-1α influenced the expression of miR-212.
Figure 4.
HIF-1α influences the expression of miR-212. miR-212 and HIF-1α mRNA expression was measured by reverse transcription-polymerase chain reaction subsequent to transfection with a siHIF-1α and HIF-1α plasmid in (A) MiaPaca2 and (B) AsPc1 cell lines. HIF-1α protein expression was measured by western blot analysis subsequent to transfection with a siHIF-1α and HIF-1α plasmid in (C) MiaPaca2 and (D) AsPc1 cell lines. *P<0.05. miR, microRNA; HIF-1α, hypoxia-inducible factor-1α; siHIF-1α, small interfering RNA targeting HIF-1α; NC, negative control.
HIF-1α directly regulates the expression of miR-212 in PDAC by binding to the HRE at the miR-212 gene promoter
The above results indicated that HIF-1α may serve a role in modulating miR-212 expression; therefore, the present study further investigated whether HIF-1α directly targeted miR-212. To demonstrate the binding of HIF-1α to the miR-212 promoter in pancreatic cancer cells, a chromatin immunoprecipitation assay was performed in MiaPaca2 cells at 1.5% O2 or 21% O2. In chromatin fractions pulled-down by an anti-HIF-1α antibody, only the HRE of the miR-212 promoter located at −481 bp was detected (Fig. 5A). The binding affinity markedly increased under hypoxic conditions compared with normal conditions.
Figure 5.
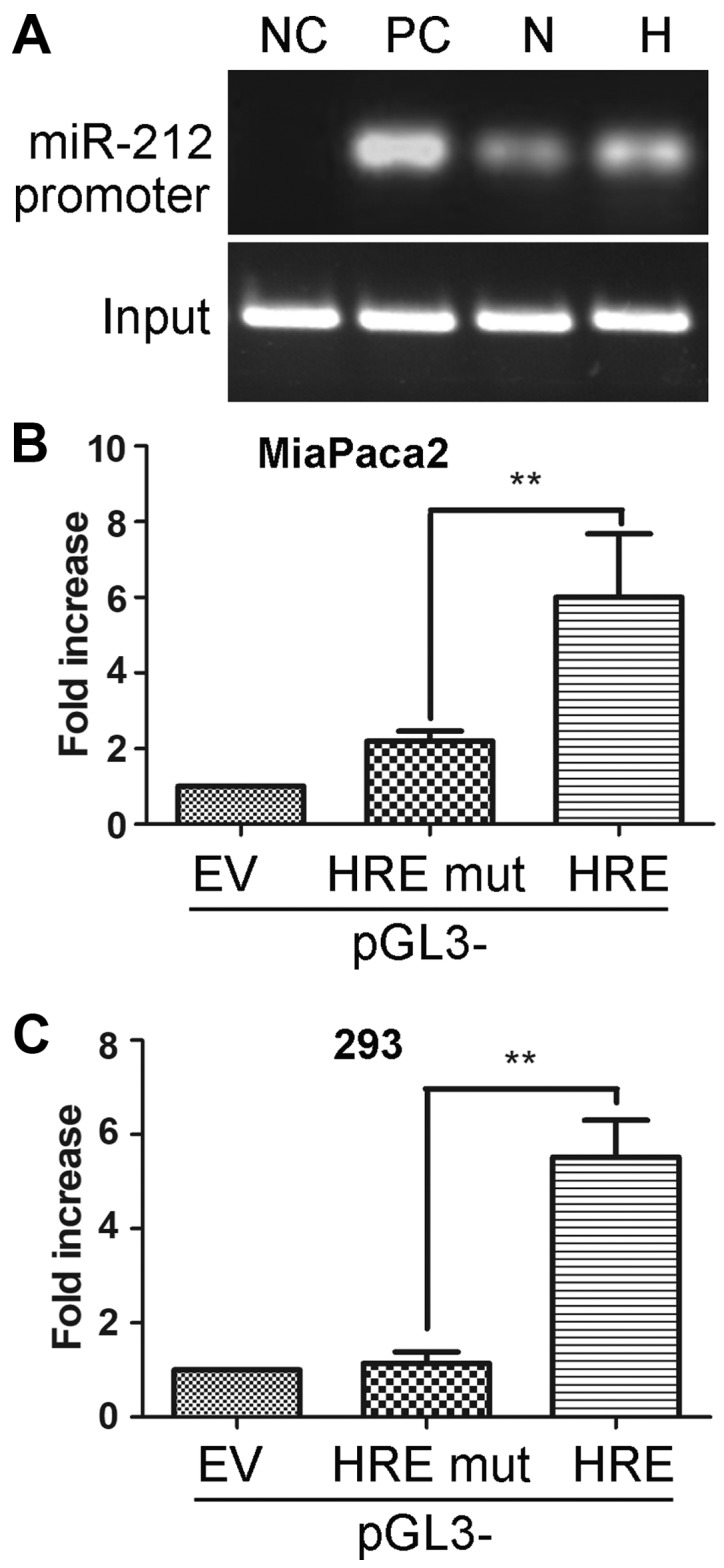
HIF-1α directly regulates the expression of miR-212 in pancreatic ductal adenocarcinoma by binding to the HRE at the miR-212 gene promoter. (A) Chromatin immunoprecipitation analysis identified HIF-1α binding to the miR-212 promoter (HRE, −481 bp) in the MiaPaca2 cell line in normoxic and hypoxic conditions. Luciferase analysis demonstrated that HIF-1α overexpression increased miR-212 promoter activity (containing HRE, −481 bp) in (B) MiaPaca2 and (C) 293 cell lines. **P<0.01. miR, microRNA; HIF-1α, hypoxia-inducible factor-1α; HRE, hypoxia response element; EV, empty vector; NC, negative control; PC, positive control; N, normoxia, H, hypoxia; mut, mutant.
To determine whether HIF-1α binding to the miR-212 promoter activated its transcription, miR-212 luciferase promoter vectors were constructed and transfected with or without a HIF-1α expression vector (pCDH-HIF-1α) into MiaPaca2 and 293 cell lines. Luciferase analysis demonstrated that HIF-1α overexpression significantly increased miR-212 promoter activity in HRE −481 bp in MiaPaca2 and 293 cell lines compared with the EV group (P<0.01; Fig. 5B and C). Additionally, the miR-212 promoter activity was not induced in the pGL3-HRE mut group. Altogether, these results suggested that HIF-1α not only directly bound to the miR-212 promoter but also upregulated the activity of miR-212 in PDACs.
Discussion
The present study investigated the association between the expression levels of miR-212 and HIF-1α in PDAC. The results revealed that miR-212 and HIF-1α mRNA expression levels were higher in PDAC samples compared with in adjacent normal pancreatic tissues. miR-212 has been demonstrated to be upregulated and downregulated in different types of solid tumors, as an oncogene and suppressor gene, respectively (17). Using clinical data analysis, associations were identified between the expression levels of miR-212 and tumor size, lymph node metastasis and vessel invasion among patients with PDAC. Notably, patients with PDAC with high miR-212 expression had significantly worse overall survival time compared with patients with low expression. The aforementioned data demonstrated that miR-212 was overexpressed in PDAC samples, and, as an oncogene, influenced the overall survival of patients with PDAC.
PDAC is characterized by stroma fibrosis, named desmoplasia, which hampers the efficacy of therapeutic treatments for PDAC (26). Desmoplasia causes hypovascularity and poor blood supply in pancreatic cancer tissues resulting in a hypoxic milieu (27). In a hypoxic microenvironment, PDAC cells constitutively express HIF-1α (28). HIF-1α as a nuclear factor may directly bind to the HREs of target genes and regulate their transcription (29). Clinical data indicated that the expression of miR-212 was correlated with the expression of HIF-1α mRNA in PDAC tissues. The expression levels of miR-212 and HIF-1α mRNA were markedly increased in hypoxic compared with normoxic conditions in vitro. In order to demonstrate the effect of HIF-1α on miR-212 expression, a HIF-1α overexpression plasmid and siHIF-1α were transfected into PDAC cell lines. The results demonstrated that miR-212 expression was upregulated subsequent to HIF-1α overexpression. By contrast, miR-212 expression was downregulated following transfection with siHIF-1α. These results revealed that there was a positive association between the expression of miR-212 and HIF-1α mRNA.
To confirm that HIF-1α regulated miR-212 expression, chromatin immunoprecipitation and dual-luciferase assays were performed. The results revealed that HIF-1α was able to directly bind to the HRE (−481 bp) of the miR-212 promoter and the binding affinity increased under hypoxic conditions. Notably, the overexpression of HIF-1α increased miR-212 promoter activity in PDAC and 293 cell lines.
In conclusion, the present study confirmed that miR-212 was upregulated in PDAC samples, compared with normal adjacent pancreatic tissues, and was associated with a poor clinical prognosis. Furthermore, a positive association between miR-212 and HIF-1α mRNA expression was identified in PDAC tissues and in vitro experiments. The results revealed that HIF-1α regulated miR-212 expression by binding to the HRE. Therefore, miR-212 may function as a novel biomarker for diagnosis, and the inhibition of miR-212 expression may be an effective therapeutic method for the treatment of patients with PDAC.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Tianjin Science and Technology Commission Joint Program (grant no. 15JCYBJC49600).
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
ZS was responsible for conception, design, study supervision, and manuscript writing, review and revision. ZS, HY and LL were responsible for the development of methodology, and acquisition, analysis and interpretation of data.
Ethics approval and consent to participate
Ethics approval was obtained from the Ethics Committee of the Tianjin Cancer Institute and Hospital (Tianjin, China). Written informed consent was obtained from individuals or their guardians.
Patient consent for publication
Patient consent for publication was obtained.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Rahib L, Smith BD, Aizenberg R, Rosenzweig AB, Fleshman JM, Matrisian LM. Projecting cancer incidence and deaths to 2030: The unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res. 2014;74:2913–2921. doi: 10.1158/0008-5472.CAN-14-0155. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 3.Adamska A, Domenichini A, Falasca M. Pancreatic ductal adenocarcinoma: Current and evolving therapies. Int J Mol Sci. 2017;18:E1338. doi: 10.3390/ijms18071338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lewis AR, Pihlak R, McNamara MG. The importance of quality-of-life management in patients with advanced pancreatic ductal adenocarcinoma. Curr Probl Cancer. 2018;42:26–39. doi: 10.1016/j.currproblcancer.2018.01.013. [DOI] [PubMed] [Google Scholar]
- 5.Uddin A, Chakraborty S. Role of miRNAs in lung cancer. J Cell Physiol. 2018 Apr 20; doi: 10.1002/jcp.26607. (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 6.Hwang HW, Mendell JT. MicroRNAs in cell proliferation, cell death, and tumorigenesis. Br J Cancer. 2006;94:776–780. doi: 10.1038/sj.bjc.6603023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang Y, Bao W, Liu Y, Wang S, Xu S, Li X, Li Y, Wu S. miR-98-5p contributes to cisplatin resistance in epithelial ovarian cancer by suppressing miR-152 biogenesis via targeting Dicer1. Cell Death Dis. 2018;9:447. doi: 10.1038/s41419-018-0390-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Idichi T, Seki N, Kurahara H, Fukuhisa H, Toda H, Shimonosono M, Okato A, Arai T, Kita Y, Mataki Y, et al. Molecular pathogenesis of pancreatic ductal adenocarcinoma: Impact of passenger strand of pre-miR-148a on gene regulation. Cancer Sci. 2018;109:2013–2026. doi: 10.1111/cas.13610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu B, Yang H, Taher L, Denz A, Grützmann R, Pilarsky C, Weber GF. Identification of prognostic biomarkers by combined mRNA and miRNA expression microarray analysis in pancreatic cancer. Transl Oncol. 2018;11:700–714. doi: 10.1016/j.tranon.2018.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xiong G, Huang H, Feng M, Yang G, Zheng S, You L, Zheng L, Hu Y, Zhang T, Zhao Y. MiR-10a-5p targets TFAP2C to promote gemcitabine resistance in pancreatic ductal adenocarcinoma. J Exp Clin Cancer Res. 2018;37:76. doi: 10.1186/s13046-018-0739-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu YA, Zhang Y, Zheng Z, Li K, Wu XH, Du QG, Ye X, Wang L, Zhu L. MicroRNA-216b reduces growth, migration and invasion of pancreatic ductal adenocarcinoma cells by directly targeting ρ-associated coiled-coil containing protein kinase 1. Oncol Lett. 2018;15:6745–6751. doi: 10.3892/ol.2018.8109. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 12.Sun D, Wang X, Sui G, Chen S, Yu M, Zhang P. Downregulation of miR-374b-5p promotes chemotherapeutic resistance in pancreatic cancer by upregulating multiple anti-apoptotic proteins. Int J Oncol. 2018 Mar 14; doi: 10.3892/ijo.2018.4315. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xie F, Huang Q, Liu CH, Lin XS, Liu Z, Liu LL, Huang DW, Zhou HC. MiR-1271 negatively regulates AKT/MTOR signaling and promotes apoptosis via targeting PDK1 in pancreatic cancer. Eur Rev Med Pharmacol Sci. 2018;22:678–686. doi: 10.26355/eurrev_201802_14293. [DOI] [PubMed] [Google Scholar]
- 14.Zhu W, Wang Y, Zhang D, Yu X, Leng X. MiR-7-5p functions as a tumor suppressor by targeting SOX18 in pancreatic ductal adenocarcinoma. Biochem Biophys Res Commun. 2018;497:963–970. doi: 10.1016/j.bbrc.2018.02.005. [DOI] [PubMed] [Google Scholar]
- 15.Yuan Q, Zhang Y, Li J, Cao G, Yang W. High expression of microRNA-4295 contributes to cell proliferation and invasion of pancreatic ductal adenocarcinoma by the down-regulation of Glypican-5. Biochem Biophys Res Commun. 2018;497:73–79. doi: 10.1016/j.bbrc.2018.02.023. [DOI] [PubMed] [Google Scholar]
- 16.Xia D, Li X, Niu Q, Liu X, Xu W, Ma C, Gu H, Liu Z, Shi L, Tian X, Chen X, Zhang Y. MicroRNA-185 suppresses pancreatic cell proliferation by targeting transcriptional coactivator with PDZ-binding motif in pancreatic cancer. Exp Ther Med. 2018;15:657–666. doi: 10.3892/etm.2017.5447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wanet A, Tacheny A, Arnould T, Renard P. miR-212/132 expression and functions: Within and beyond the neuronal compartment. Nucleic Acids Res. 2012;40:4742–4753. doi: 10.1093/nar/gks151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xie M, Fu Z, Cao J, Liu Y, Wu J, Li Q, Chen Y. MicroRNA-132 and microRNA-212 mediate doxorubicin resistance by down-regulating the PTEN-AKT/NF-κB signaling pathway in breast cancer. Biomed Pharmacother. 2018;102:286–294. doi: 10.1016/j.biopha.2018.03.088. [DOI] [PubMed] [Google Scholar]
- 19.Wu Z, Zhou L, Ding G, Cao L. Overexpressions of miR-212 are associated with poor prognosis of patients with pancreatic ductal adenocarcinoma. Cancer Biomark. 2017;18:35–39. doi: 10.3233/CBM-160671. [DOI] [PubMed] [Google Scholar]
- 20.Jia P, Wei G, Zhou C, Gao Q, Wu Y, Sun X, Li X. Upregulation of MiR-212 inhibits migration and tumorigenicity and inactivates Wnt/β-catenin signaling in human hepatocellular carcinoma. Technol Cancer Res Treat. 2018;17:1533034618765221. doi: 10.1177/1533034618765221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hu B, Jin X, Wang J. MicroRNA-212 targets mitogen-activated protein kinase 1 to inhibit proliferation and invasion of prostate cancer cells. Oncol Res. 2018;26:1093–1102. doi: 10.3727/096504018X15154112497142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tang T, Huan L, Zhang S, Zhou H, Gu L, Chen X, Zhang L. MicroRNA-212 functions as a tumor-suppressor in human non-small cell lung cancer by targeting SOX4. Oncol Rep. 2017;38:2243–2250. doi: 10.3892/or.2017.5885. [DOI] [PubMed] [Google Scholar]
- 23.Jiang C, Wang H, Zhou L, Jiang T, Xu Y, Xia L. MicroRNA-212 inhibits the metastasis of nasopharyngeal carcinoma by targeting SOX4. Oncol Rep. 2017;38:82–88. doi: 10.3892/or.2017.5641. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 24.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 25.Zhao T, Jiang W, Wang X, Wang H, Zheng C, Li Y, Sun Y, Huang C, Han ZB, Yang S, et al. ESE3 inhibits pancreatic cancer metastasis by upregulating E-cadherin. Cancer Res. 2017;77:874–885. doi: 10.1158/0008-5472.CAN-16-2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Whatcott CJ, Diep CH, Jiang P, Watanabe A, LoBello J, Sima C, Hostetter G, Shepard HM, Von Hoff DD, Han H. Desmoplasia in primary tumors and metastatic lesions of pancreatic cancer. Clin Cancer Res. 2015;21:3561–3568. doi: 10.1158/1078-0432.CCR-14-1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu H, Ma Q, Xu Q, Lei J, Li X, Wang Z, Wu E. Therapeutic potential of perineural invasion, hypoxia and desmoplasia in pancreatic cancer. Curr Pharm Des. 2012;18:2395–2403. doi: 10.2174/13816128112092395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Akakura N, Kobayashi M, Horiuchi I, Suzuki A, Wang J, Chen J, Niizeki H, Kawamura Ki, Hosokawa M, Asaka M. Constitutive expression of hypoxia-inducible factor-1alpha renders pancreatic cancer cells resistant to apoptosis induced by hypoxia and nutrient deprivation. Cancer Res. 2001;61:6548–6554. [PubMed] [Google Scholar]
- 29.Dengler VL, Galbraith M, Espinosa JM. Transcriptional regulation by hypoxia inducible factors. Crit Rev Biochem Mol Biol. 2014;49:1–15. doi: 10.3109/10409238.2013.838205. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.