Figure 3. uS10 in colliding ribosome is efficiently ubiquitinated.
-
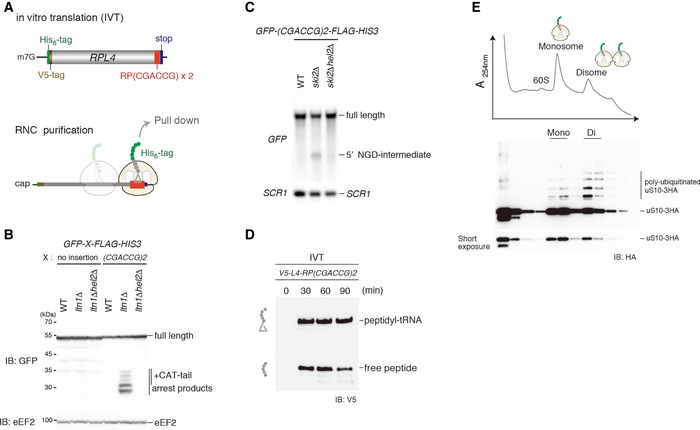
ATop: schematic drawing of the (CGA‐CCG) dicodon reporter mRNA used for in vitro translation experiments. Bottom: scheme outlining the principle of ribosome–nascent chain (RNC) purification. Stalled and colliding ribosomes are pulled down via an affinity tag on the nascent chain.
-
B, CBoth RQC and NGDRQC+ are triggered by a (CGA‐CCG) dicodon containing arrest sequence in vivo. (B) Western blot showing the arrest products derived from the GFP‐X‐FLAG‐HIS3 reporter. Translation products were detected by Western blot using an anti‐GFP antibody. (C) Northern blot for the 5′ NGD‐IM derived from the (CGA‐CCG) reporter in ski2Δ cells. 5′ NGD‐IMs were detected with a DIG‐labelled GFP probe. SCR1 was used as a load control.
-
DWestern blot of test translations using the (CGA‐CCG) dicodon stalling mRNA reporter shown in (A). The mRNA reporter was added to a yeast in vitro translation extract obtained from a ski2ΔuS10‐3HA strain. Expression of the translation products (free His‐ and V5‐tagged truncated uL4 protein and the same protein attached to tRNA) was visualized with an anti‐V5 antibody.
-
ESucrose gradient fractions (top) and Western blot analysis (bottom) of the (CGA‐CCG) dicodon‐stalled RNC samples. After the translation reaction, the RNCs were affinity‐purified as indicated in (C). The eluate was loaded on a 10–50% sucrose gradient and fractionated. Each collected fraction was analysed using anti‐HA antibody to detect uS10‐HA. Note that disomes are preferentially polyubiquitinated over monosomes.
