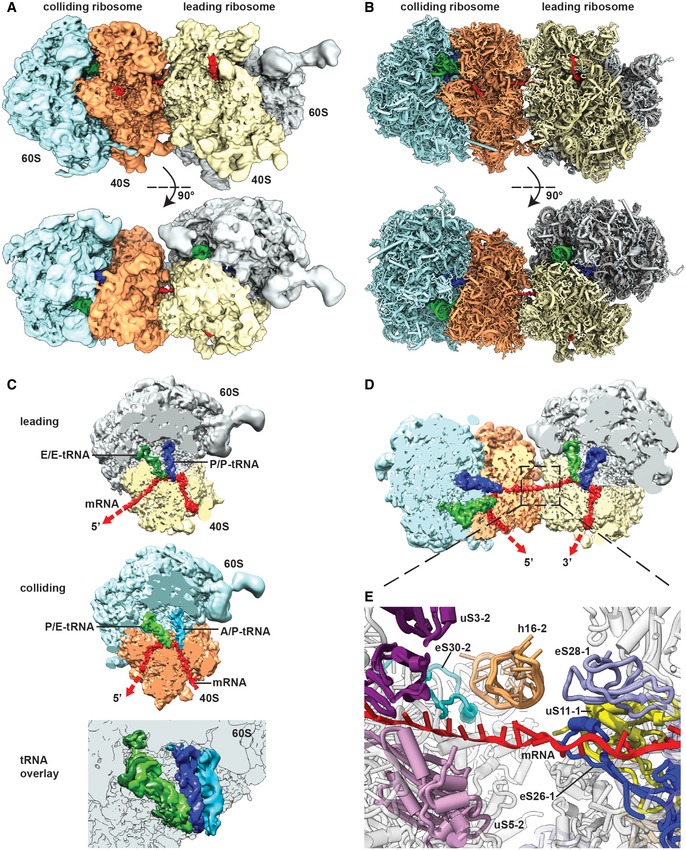
Figure 4. Cryo‐EM structure and molecular model of a stalled disome.
-
A, BSide view and top view of the Cryo‐EM reconstruction (A) and molecular model (B) of the disome stalled on a (CGA‐CCG) reporter mRNA. The first ribosome in the disome stalled on the dicodon mRNA is referred to as the leading ribosome and the second as the colliding ribosome.
-
CCut top views on the leading and colliding ribosome. The leading ribosome is in a non‐rotated POST‐state containing tRNAs in P/P and E/E states and an empty A‐site, whereas the colliding ribosome is in a rotated state containing hybrid A/P and P‐/E‐site tRNAs. An overlay is shown omitting the 40S subunit for clarity.
-
DCut top view on the disome with focus on the mRNA density highlighted in red.
-
EZoom on the mRNA at the interface of the leading and colliding ribosome. The mRNA exits the leading ribosome close to ribosomal proteins uS11, eS26 and eS28 and enters the colliding ribosome near h16, uS3, uS5 and eS30.
