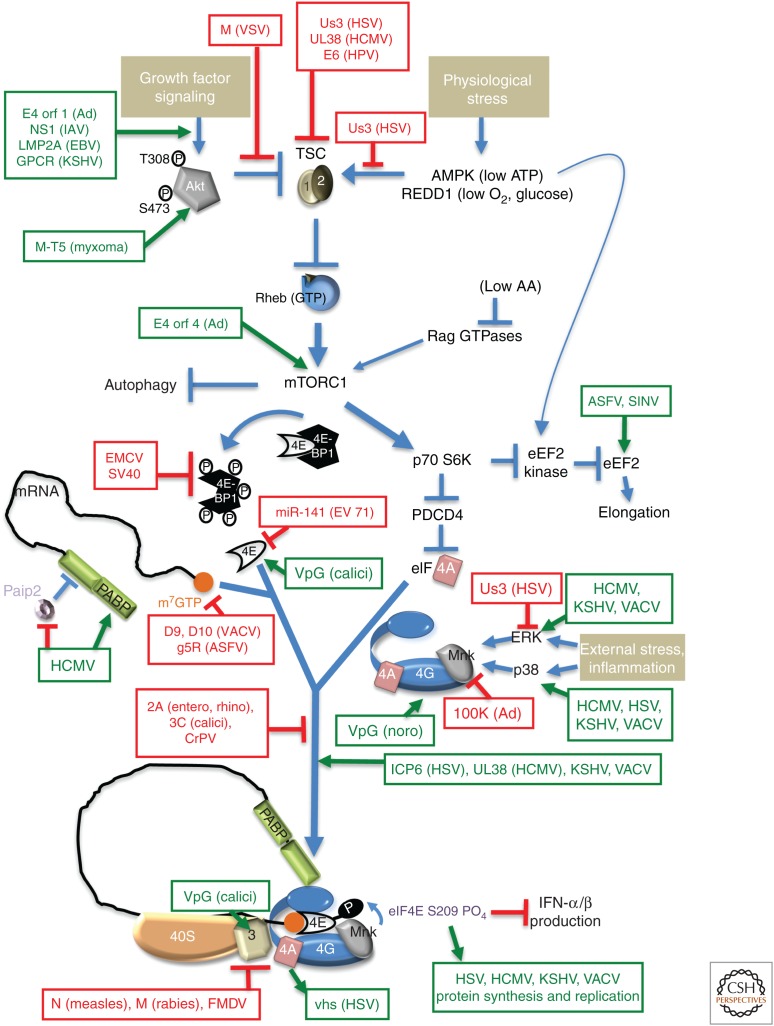
Figure 3.
Subverting mechanistic target of rapamycin complex 1 (mTORC1) signaling to control translation in virus-infected cells. Occupying a nexus of intersecting signaling networks, the cellular ser/thr kinase mTORC1 plays a critical role stimulating anabolic programs, like protein synthesis, and repressing catabolic outcomes like autophagy. Briefly, growth factor–induced Akt phosphorylation (T308, S473) and activation represses TSC1/2, which, in turn, allows Rheb-GTP to activate mTORC1. Nutrient, energy, amino acid (aa), or oxygen insufficiency (physiological stress) all repress mTORC1 through discrete effectors. Signaling through mTORC1 allows swift changes in translational output in response to differing environmental and physiological inputs by controlling initiation and elongation. Initiation is stimulated by phosphorylating and inactivating 4E-BP translational repressor family members (e.g., 4E-BP1), which bind the cap-binding protein eIF4E to prevent its interaction with eIF4G. Through its substrate p70S6K, mTORC1 controls the DEAD-box-containing RNA helicase eIF4A, which together with eukaryotic initiation factor (eIF)4E and eIF4G comprises the multisubunit initiation factor eIF4F. Regulated eIF4F assembly controls cap-dependent mRNA translation as eIF4F recruits 40S subunits to the mRNA capped 5′ end. By repressing eEF2 kinase, mTORC1 stimulates eEF2 and elongation. The impact of mTORC1 activation on translation initiation and elongation is shown, as are viral factors that stimulate (green) and repress (red) the indicated cellular effectors. SV40, simian virus 40; calici, calicivirus; noro, norovirus; entero, enterovirus; rhino, rhinovirus.