Figure 4.
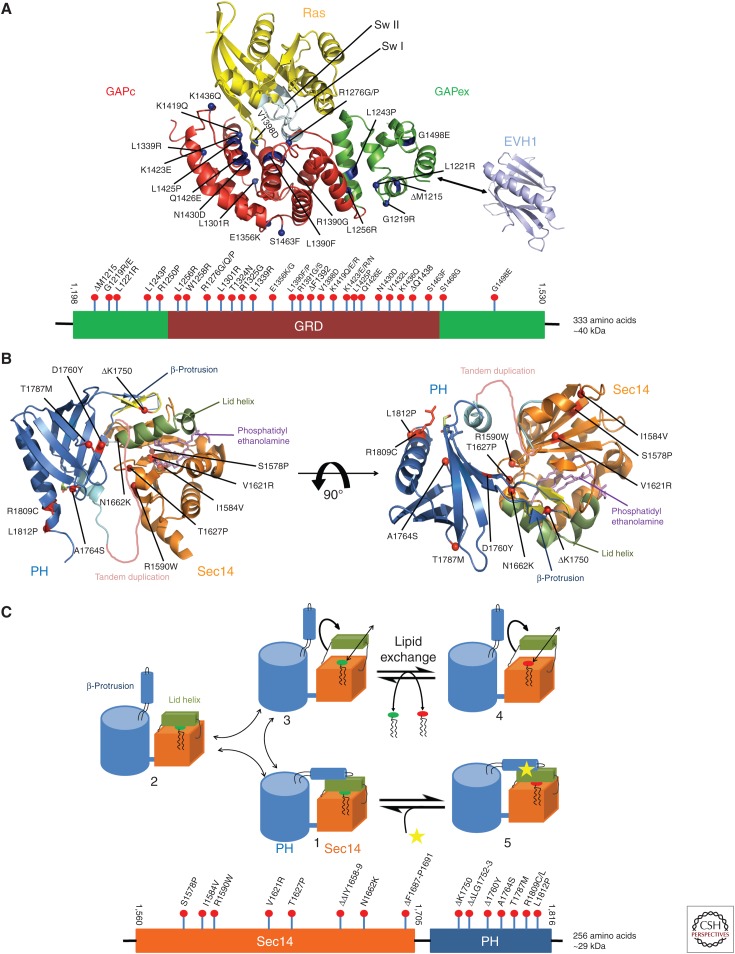
Structural components of neurofibromin. (A) Ribbon representation of a putative Ras–GRD complex, modeled on the basis of the GAP-related domain (GRD) structure (Scheffzek et al. 1998a) and the Ras-p120GAP complex (Scheffzek et al. 1997a). Blue dots indicate the location of missense mutations found in neurofibromatosis type 1 (NF1) patients. The EVH1 domain of Spred1 shown to interact with extra domain (GAPex) is depicted in cyan. Below is the domain scheme of GRD of neurofibromin with amino-terminal and carboxy-terminal extra domains with nontruncating patient-derived mutations included (see text). (B) Ribbon diagram of the Sec14-PH module bound to lipid with approximately 90° rotation about a horizontal axis. Orange ribbon represents Sec14-like domain bound to lipid (phosphatidyl ethanolamine) (in violet). Pleckstrin homology (PH)-like domain is shown in blue. Helix represented in green is a part of Sec14 that forms lid helix. Red dots indicate the location of missense mutations found in NF1 patients. A tandem duplication and ΔK1750 are also shown. Below is the domain scheme of Sec14-PH of neurofibromin with nontruncating mutations identified in NF1 patients (see text). (C) Cartoon of the Sec14-PH module, illustrating the proposed structural changes associated with the exchange of glycerophospholipid ligands. Starting from the observed structure (1), lid helix (in green), and β-protrusion helix can move either stepwise (1 > 2 > 3) or in a concerted fashion (1 > 3) into an open conformation, which allows the exchange of lipid molecules between Sec14-PH and membrane (4). Binding of modulator to the lid helix or the β-protrusion appears to prevent the lipid exchange reaction (5). (Structural visualizations in panels A and B were done with The PyMOL Molecular Graphics System, Version 1.8 Schrödinger, LLC.)