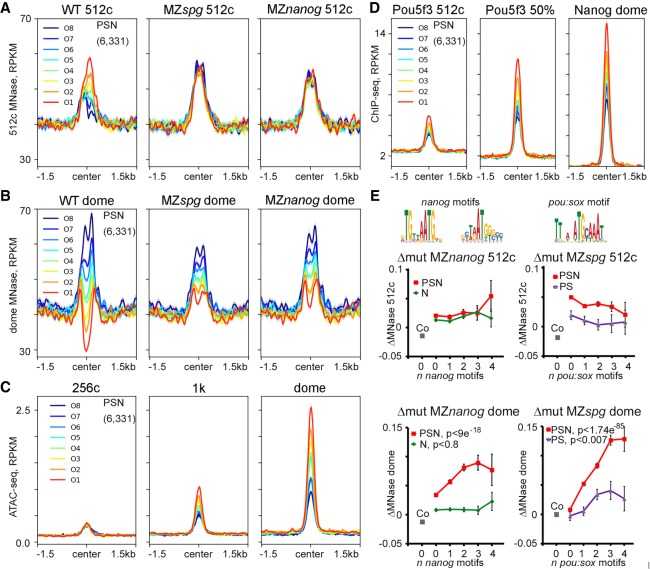
Figure 2.
Pou5f3 and Nanog displace nucleosomes from different regions before and after ZGA. (A–D) PSN group regions ranked into octiles by ascending difference between WT post-ZGA and pre-ZGA MNase-seq signal (ΔWT post-pre). Three-kilobase genomic regions were aligned at the ChIP-seq peak centers. Summary plots per octile: (A) nucleosome occupancy 512-cell stage, (B) nucleosome occupancy dome, (C) ATAC-seq, and (D) TF occupancy. (E) TF effects on nucleosome density were estimated as MNase-seq signal difference between the WT and indicated mutants (Δmut) and related to the number of specific motifs (zero, one, two, three, or four or more motifs) per 320-bp binding region. Motifs are indicated on top. Pou5f3 and Nanog act nonsequence-specifically at the 512-cell stage (upper row). At dome stage (lower row), Pou5f3 and Nanog act by sequence-specific binding to their motifs on triple-occupancy PSN regions but not on single and double TFBSs. PSN indicated by red; N, green; and PS, violet. (Co) Random control regions (zero motifs; gray); (y-axis) Δmut.