Figure 1.
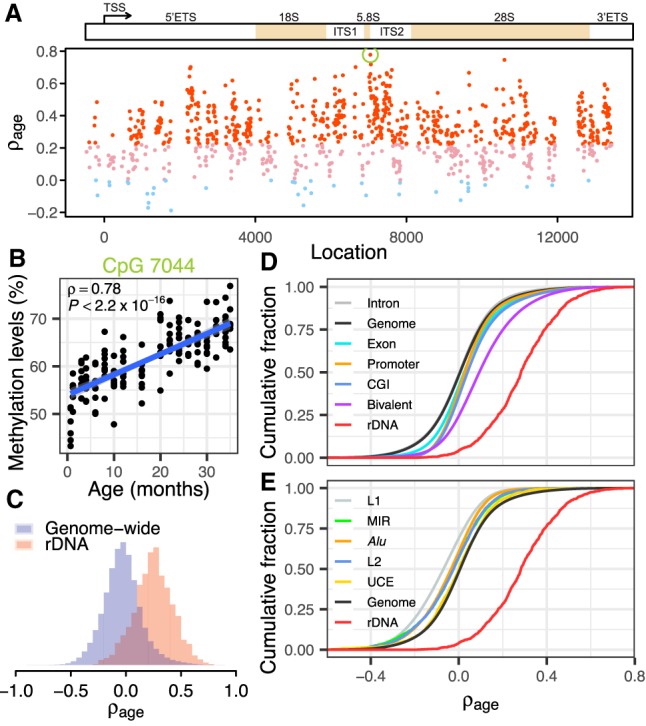
Ribosomal DNA methylation is strongly associated with age. (A) Age-associated hypermethylation of rDNA CpGs in mice. Spearman's correlation coefficients with age (ρage) for CpGs along the rDNA sequence. Red dots indicate CpGs with significant positive correlation with age (ρage > 0, FDR < 0.01); pink and light blue dots denote nonsignificant CpGs (FDR > 0.01) with positive and negative coefficients, respectively. The green circle indicates CpG 7044. (B) Scatterplot shows the correlation between CpG 7044 methylation and age (ρ = 0.78, P < 2.2 × 10−16). (C) The distribution of ρage for CpGs within the rDNA and across the whole genome (Wilcoxon rank-sum test, P < 2.2 × 10−16). (D,E) Cumulative distribution of correlation coefficients for various genomic elements. CpGs within CpG islands (CGIs) and bivalent chromatin have significantly higher ρage than the genome-wide background, although both of them are significantly lower than rDNA CpGs (P < 2.2 × 10−16). Repetitive elements, including L1, L2, Alu (B elements), and mammalian-wide interspersed repeats (MIR), tend to be hypomethylated during aging (P < 2.2 × 10−16) relative to the genome-wide background. All features on D, except for intron, show a shift to the right of the genome-wide background. All features on E, except for the rDNA, show a shift to the left of the genome-wide background. Also see Supplemental Figure S5.
