Figure 3.
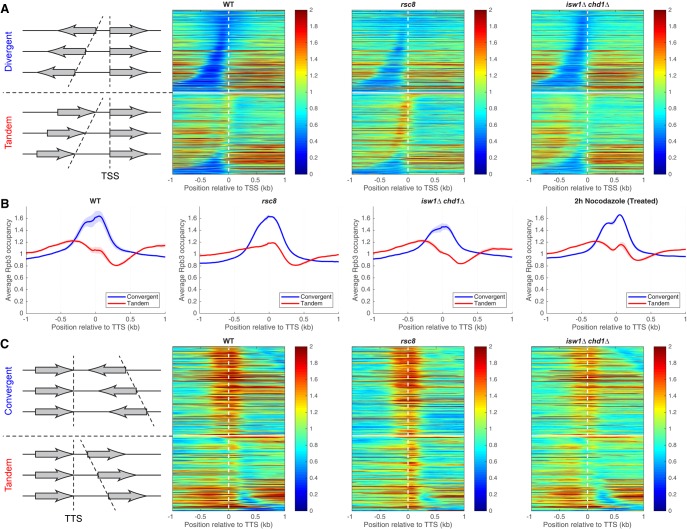
RSC and ISW1/CHD1 have opposite effects on the levels of elongating and terminating Pol II. (A) Pol II occupancy heat maps. Genes were divided into divergent (top) and tandem (bottom) pairs (separated by the white horizontal line), which were then sorted by intergenic distance and aligned on the TSS of the downstream gene (dashed vertical line). Data for wild type, rsc8, and isw1Δ chd1Δ double mutant cells. Rpb3 ChIP-seq data for DNA fragments of 300 bp or less normalized to the genomic average (set at 1). Shaded areas indicate the range of biological replicate data. (B) Aggregate plots for Pol II occupancy (IP/Input) relative to the TTS of the upstream gene for convergent and tandem gene pairs in wild type, rsc8, isw1Δ chd1Δ double mutant, and nocodazole-treated wild-type cells. (C) Heat maps for the data in B, aligned on the TTS of the upstream gene and sorted by the distance to the TSS of the downstream gene. The white horizontal line separates convergent (top) from tandem gene pairs (bottom). Equivalent plots for the other strains are shown in Supplemental Figures S5, S6.