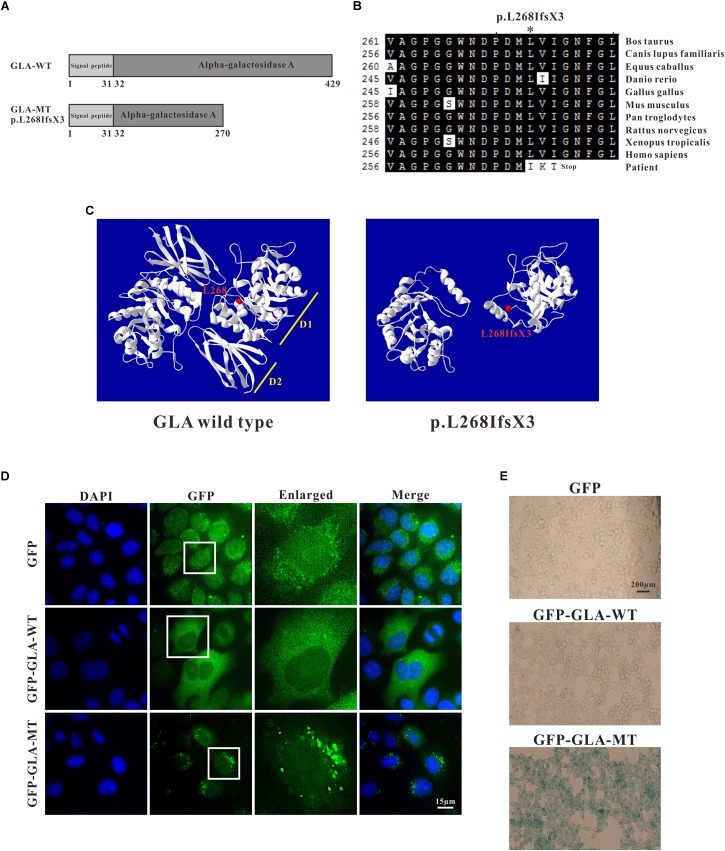
FIGURE 4.
Analysis of GLA mutation. (A) Schematics of the secondary structure and functional domains of the GLA protein. (B) Evolutionary conservation of amino acid residues altered by c.801 + 1G > A (p.L268IfsX3) across different species. NCBI accession numbers are: Bos Taurus: NP_001179665; Canis lupus familiaris: XP_538109; Equus caballus: XP_001492699; Danio rerio: NP_001006103; Gallus gallus: XP_420183; Mus musculus: NP_038491; Pan troglodytes: XP_003954083; Rattus norvegicus: NP_001102290; Xenopus tropicalis: NP_001120606; Homo sapiens: NP_000160. (C) The mutant protein p.L268IfsX3 were predicted to result in the loss of the C-terminal domain by Swiss-Model online software compared to the wild type. Ribbon representation of the human GLA and map of the studied variant localization obtained by homology modeling analysis. The WT and MT monomers are shown in white. The altered amino acid is shown as red ball. Each monomer is composed of two domains as pointed out: Domain 1 (D1) with the catalytic site, and Domain 2 (D2). (D) HEK293T were transfected with GFP alone, GFP-GLA-WT, and GFP-GLA-MT plasmids and the localization of wild type and mutant GLA were studied by immunofluorescence. Bar: 15 μm. (E) Visualization of senescence associated β-galactosidase show differences between wild type and mutant GLA transfected cells. Examination for staining was done after incubation for 4 to 8 h under bright field microscopy at 200× magnification. Bar: 200 μm.