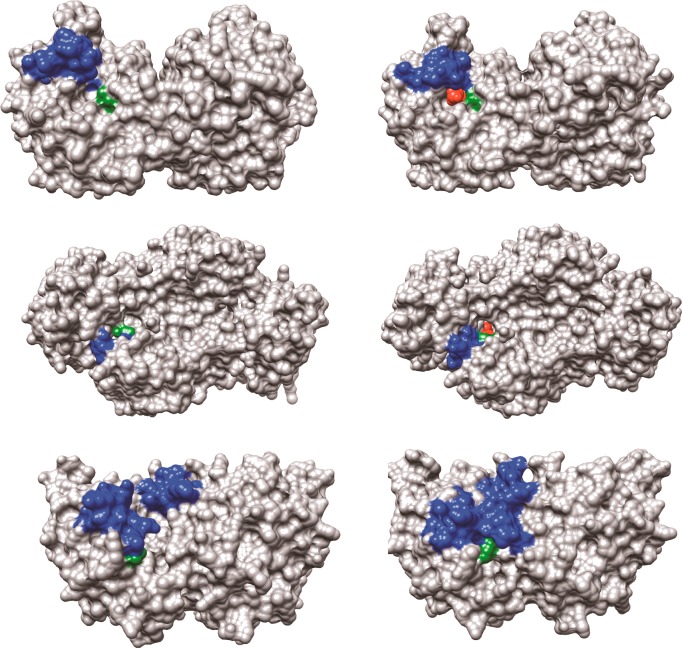
Figure 2.
Surface structures for TIM (top), GPDH (middle), and OMPDC (bottom). The binding energy of the ligand phosphodianion is utilized to immobilize these loops, in driving the conformational changes to the stiff and catalytically active closed structures shown on the right. The ligand phosphodianion at the closed enzymes is shaded red, and the side-chain cations, which interact with the phosphodianion, are shaded green. Key: Top structures; TIM from Trypanosoma brucei brucei (open form, PDB entry 3TIM; closed form with 3-phosphoglycerate bound, PDB entry 1IIH). The phosphodianion gripper loop (residues 165–177) is shaded blue, and the side chain from K12 is shaded green. Not shown is loop 7 (residues 208–216), whose side chains Y208 and S211 move as the planes defined by the peptide bonds from G209 and G210 undergo 90° and 180° rotations, respectively.74 Middle structures; GPDH from human liver (open form, PDB entry 1X0V; closed form with NAD and DHAP bound, PDB entry 1WPQ). The phosphodianion gripper loop (residues 292–297) is shaded blue, and the side chain from R269 is shaded green. The side chain of Q295 interacts with the substrate phosphodianion through the intervening side chain of R269.75 Bottom structures; OMPDC from Saccharomyces cerevisiae (open form, PDB entry 1DQW; closed form with 6-hydroxyuridine 5′-monophosphate bound, PDB entry 1DQX). The phosphodianion gripper loop (residues 202–220) is shaded blue, and the side chain from R235 is shaded green. The pyrimidine umbrella loop (residues 151–165) is also shaded blue. The blue loops interact at the closed form of OMPDC through a hydrogen bond between the side chains of S154 and Q215.76,77