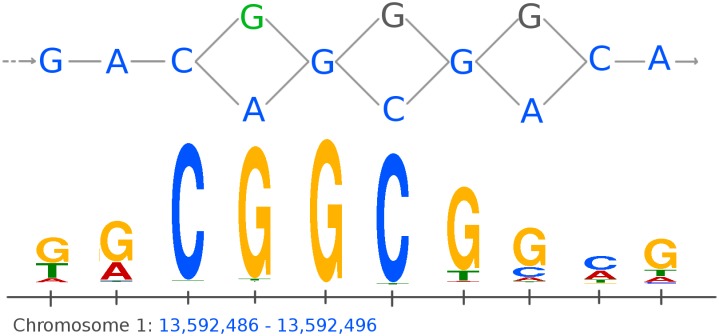
Fig 3. Example of a motif match for a peak following a variant not part of the linear reference genome.
Showing part of the graph-based reference genome for A. thaliana on chromosome 1 (top) with the linear reference genome represented by blue nodes (bases). The figure shows a peak found for the ERF115 transcription factor that matches a DNA-binding motif. The peak detected by Graph Peak Caller follows the linear reference genome (blue nodes) except for the first SNP shown (green node), where the peak follows the green G instead of the blue A, making this a significant match against the DNA-binding motif (shown as a sequence logo at the bottom). The peak detected by MACS2 does not have a significant motif match. All alignments intersecting with this binding site are present in a common haplotype among the haplotypes used to build the reference graph (S2 Appendix).