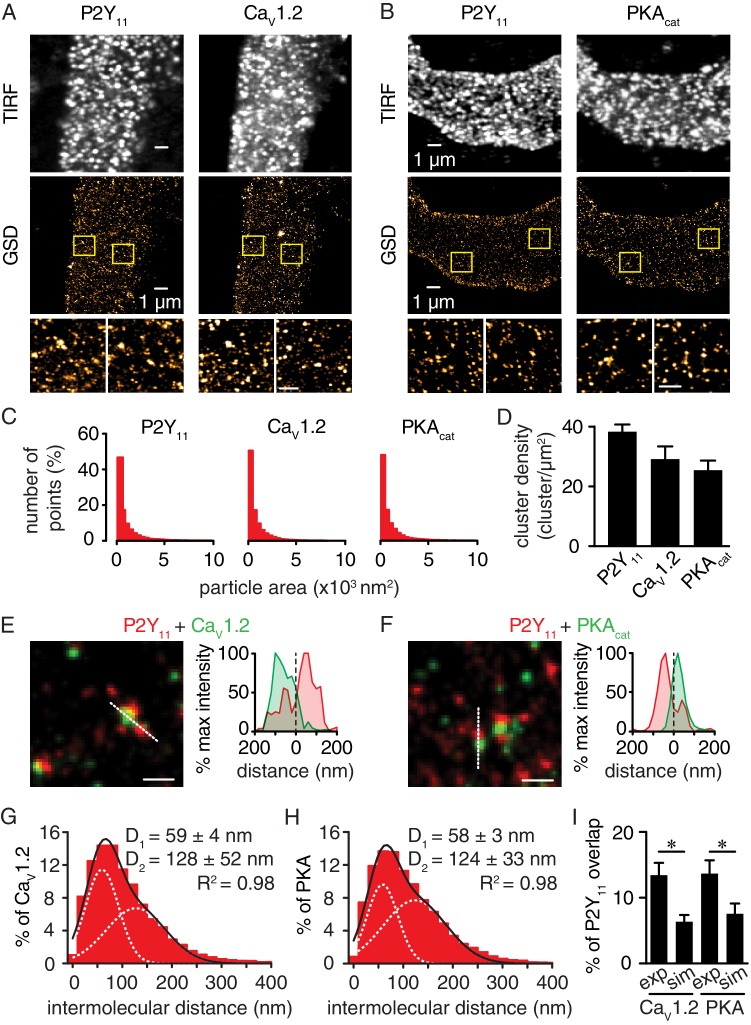
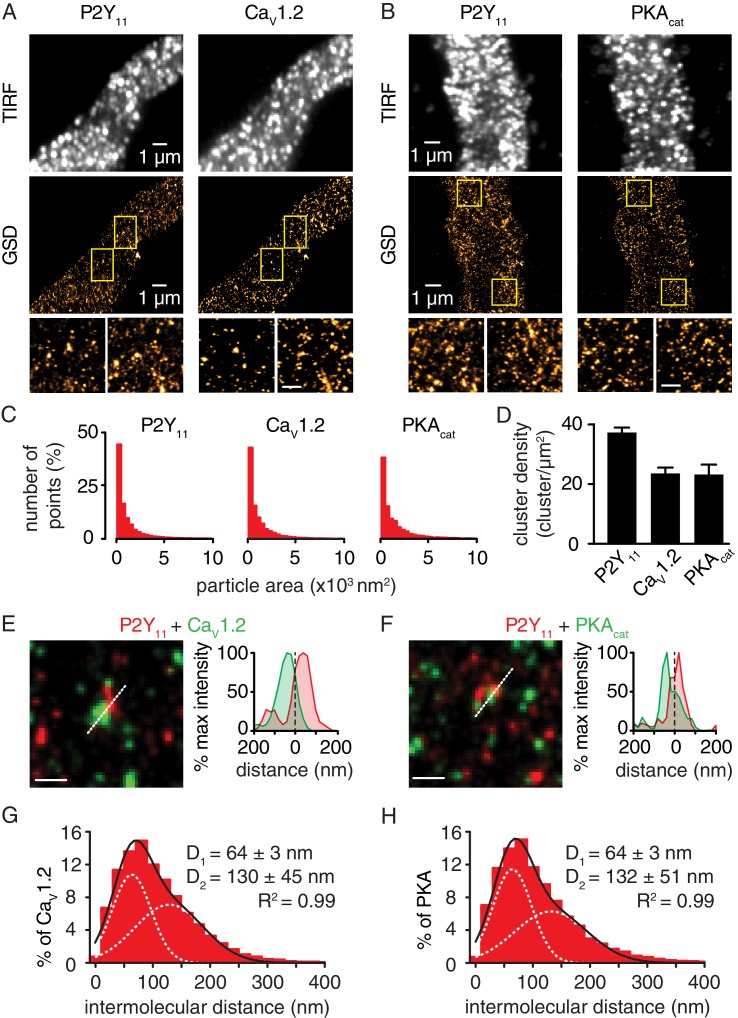
Figure 3. Nanometer organization of P2Y11 with CaV1.2 and PKAcat in human arterial myocytes.
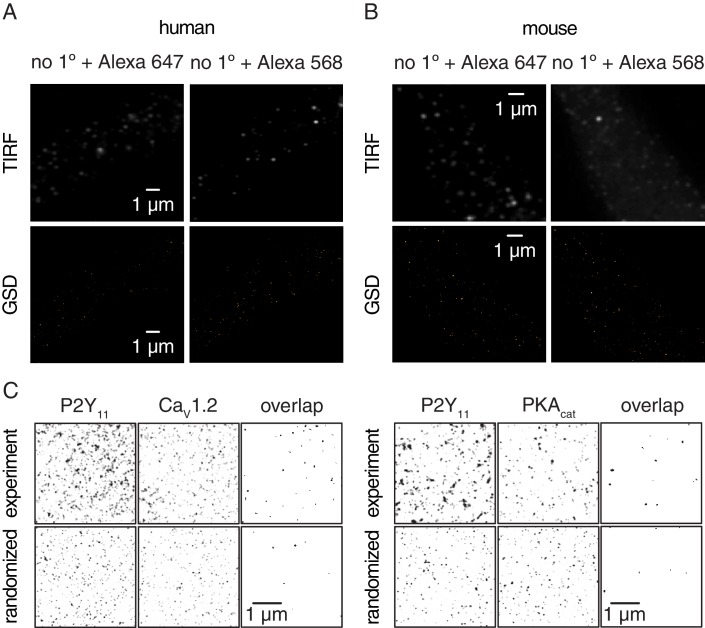
Representative conventional TIRF images (top) and corresponding GSD reconstruction maps (bottom) from human arterial myocytes labeled for (A) P2Y11 and CaV1.2 and (B) P2Y11 and PKAcat. Lower panels display enhanced magnifications of areas shown in yellow boxes (scale bar, 400 nm). (C) Histograms of the area of P2Y11, CaV1.2 and PKAcat clusters in isolated human arterial myocytes (1621 ± 29, 1209 ± 16 and 1322 ± 20 nm2, respectively; Figure 3—source data 1). (D) Bar plot of cluster density for P2Y11, CaV1.2 and PKAcat (38 ± 2, 29 ± 4, and 25 ± 3 clusters/µm2, respectively; Figure 3—source data 2). Enlarged merged image (left) and associated x-y fluorescence intensity profile (right) from area highlighted by the dotted lines of sites of close proximity between (E) P2Y11 (red) and CaV1.2 (green) and (F) P2Y11 (red) and PKAcat (green) (scale bar, 200 nm). Histograms of the lowest intermolecular distance to P2Y11 centroids for (G) CaV1.2 (n = 19,611 particles from 6 cells; Figure 3—source data 3) and (H) PKAcat (n = 22,425 particles from 6 cells; Figure 3—source data 4) fluorescence particles. Data were fit with a sum of two Gaussian functions with depicted R2 and centroids. (I) Bar plot of % overlap of P2Y11 with CaV1.2 or PKAcat for experimental (CaV1.2: n = 36 segments from 12 cells; PKAcat: n = 22 segments from 11 cells) and randomized simulations images (CaV1.2: n = 6 segments from 6 cells; PKAcat: n = 6 segments from 6 cells) (*p<0.05, unpaired t test with Welch’s correction; Figure 3—source data 5).
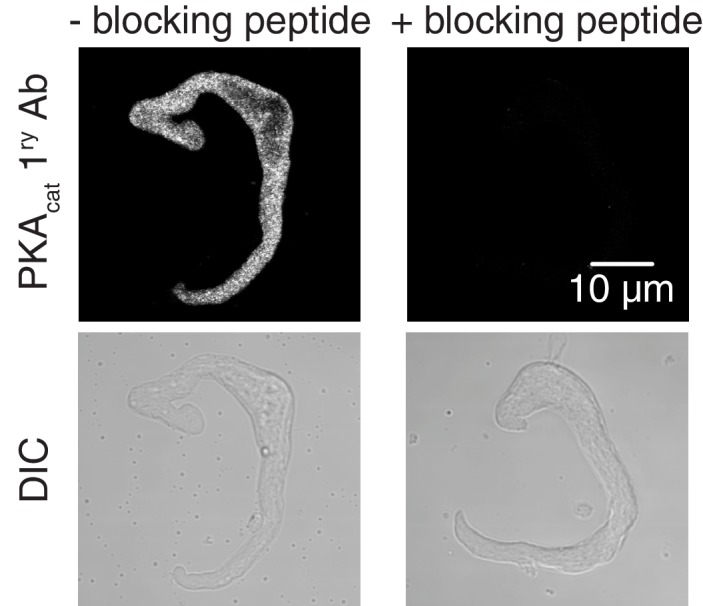
Figure 3—figure supplement 1. Validation for PKAcat primary antibody.