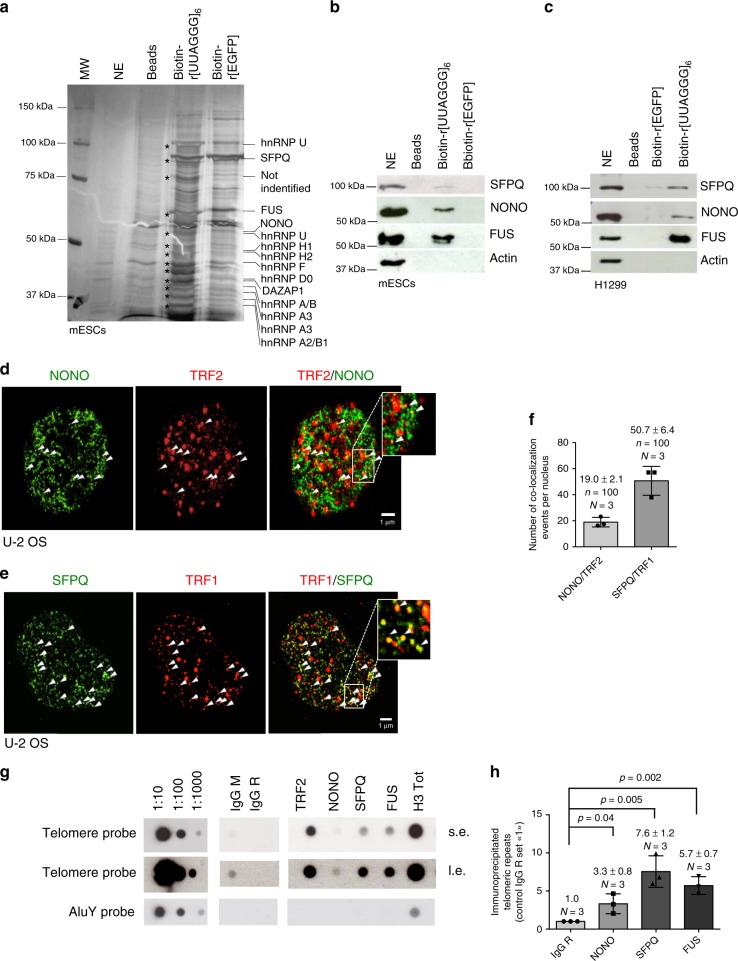
Fig. 1.
SFPQ and NONO interact with TERRA and localize to telomere repeats. a Silver staining of SDS-PAGE gels of eluates obtained from RNA-pull-down experiments using biotinylated r[UUAGGG]6, biotinylated r[EGFP] RNA oligonucleotides or empty beads. Candidate TERRA interacting proteins identified by mass spectrometry are indicated. NE, nuclear extract used as input; MW, molecular weight marker. b, c Western blotting analysis of RNA-pull-down eluates using specific anti-NONO and anti-SFPQ antibodies confirms binding specificity of NONO and SFPQ for UUAGGG RNA repeats in mESCs (b) and H1299 cells (c). FUS was previously reported to interact with TERRA42. Actin was used as loading control; NE, nuclear extract was used as input. Source blots are available as Supplementary Figure 6 and 7. d Representative image of co-localization events (arrowheads) between NONO and the shelterin protein TRF2 as determined by confocal microscopy in U-2 OS cells. e Representative image of co-localization events (arrowheads) between SFPQ and the shelterin protein TRF1 by confocal microscopy in U-2 OS cells. f Quantification of d and e. Mean number of NONO-TRF2 and SFPQ-TRF1 co-localization events per nucleus is shown. Error bars indicate standard deviation. n = number of analyzed nuclei. g Chromatin immunoprecipitation experiments (ChIP) using U-2 OS cells and mouse anti-TRF2, rabbit anti-histone H3, rabbit anti-FUS, rabbit anti-NONO, and rabbit anti-SFPQ antibodies. Mouse and rabbit control IgGs (IgG M/IgG R) were used as negative control. Serial dilutions of chromatin extract (input) prepared from U-2 OS cells were loaded. h Quantification of three independent ChIP experiments, average enrichment of telomeric repeats is indicated; s.e.: short exposure; l.e.: long exposure. f, h N, number of independent experiments, whiskers indicate standard deviation; a two-tailed Student’s t-test was used to calculate p-values. Source data is provided as a Supplementary Information File. Scale bar, 1 μm. siRNAs listed in Supplementary Table 1