Fig. 3.
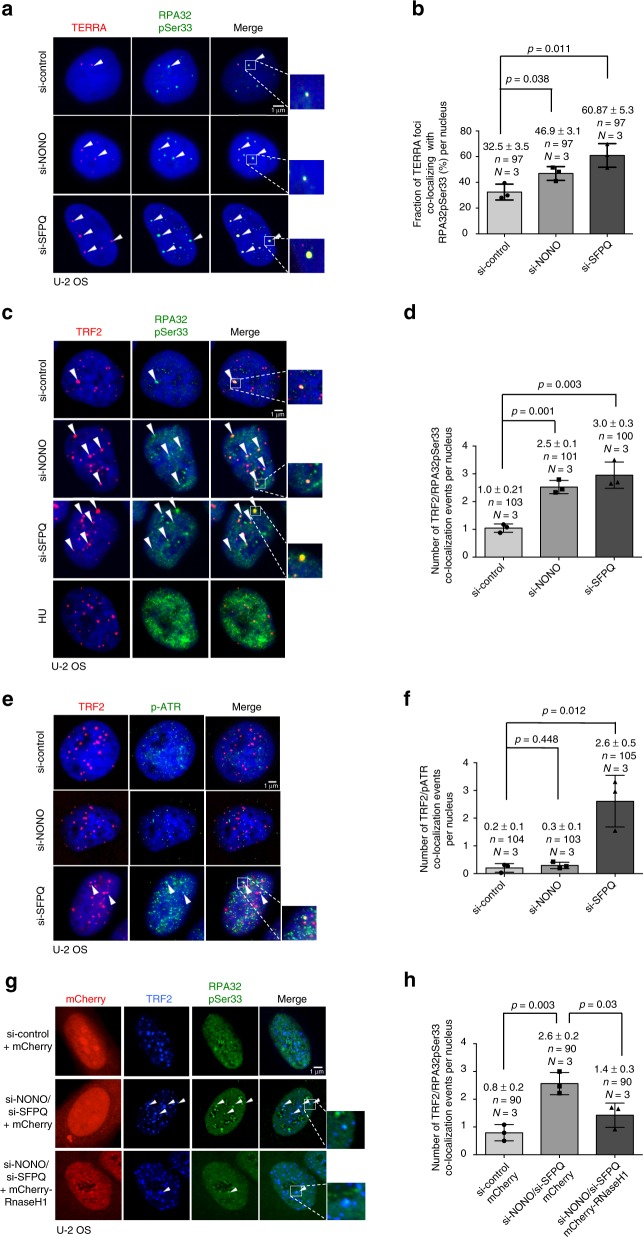
Depletion of NONO and SFPQ cause replication defects at telomeres in U-2 OS cells. a Representative images of TERRA RNA-FISH combined with anti-RPA32pSer33 immunostaining performed in U-2 OS cells transfected with indicated siRNAs. Arrowheads indicate co-localization events. b Quantification of a; percentage of TERRA foci co-localizing with RPA32pSer33 per nucleus is indicated. c Representative images of combined immunofluorescence with anti-TRF2 and anti-RPA32pSer33 antibodies in U-2 OS cells transfected with indicated siRNAs. Arrowheads indicate co-localization events. Induction of replicative stress by treating cells with hydroxyurea (5 mM) for 6 h was used as control for RPA32pSer33 staining. d Quantification of TRF2/RPApSer33 co-localization events per nucleus. e Representative images of combined immunofluorescence using anti-TRF2 and anti-pATR antibodies (S428) in U-2 OS cells transfected with indicated siRNAs. Arrowheads indicate co-localization events. f Quantification of TRF2/pATR co-localization events per nucleus. g Representative images of combined immunofluorescence using anti-TRF2 and anti-RPA32pSer33 antibodies on U-2 OS cells transfected with indicated siRNAs and transiently expressing mCherry-RNase H1 or mCherry empty vector. Arrowheads indicate co-localization events. h Quantification of the number of TRF2/RPA32pSer33 co-localizations per nucleus. For quantifications in b, d, f, h, mean values and standard deviations are reported, error bars indicated standard deviation. N = number of independent experiments. n = number of analyzed nuclei. A two-tailed Student’s t-test was used to calculate statistical significance; p-values are shown. Source data is provided as a Supplementary Information File. Scale bar, 1 μm. siRNAs listed in Supplementary Table 1