Fig. 1.
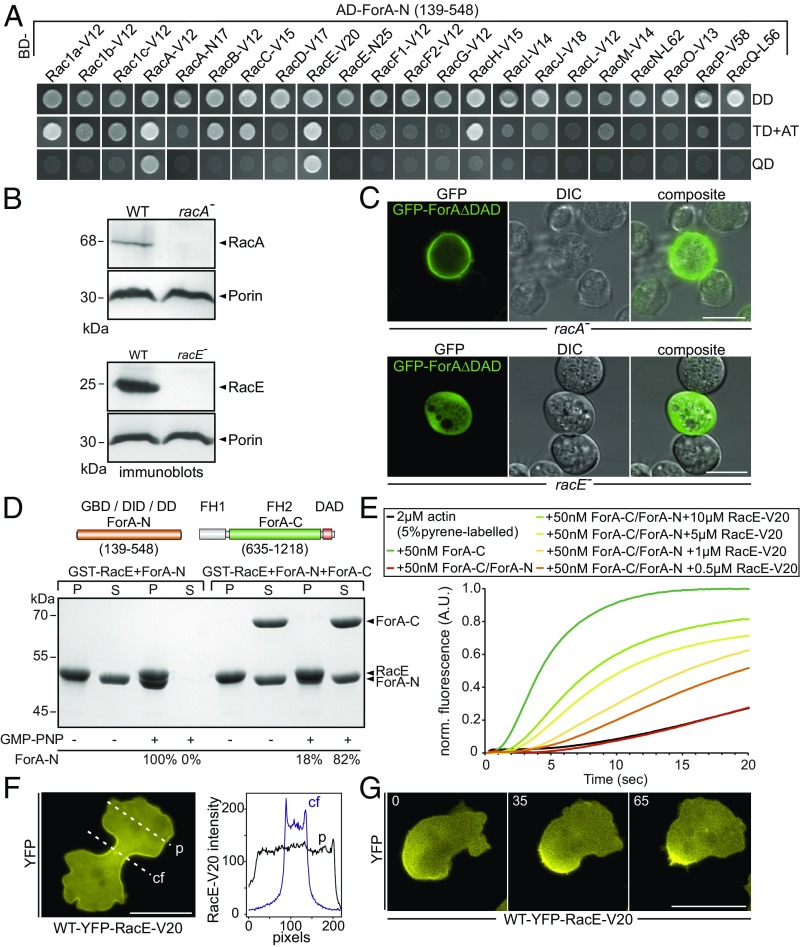
ForA is regulated by the RhoA-homolog RacE. (A) The N-terminal domain of ForA (ForA-N) containing the GBD interacts specifically with the activated forms of RacA (RacA-V12) and RacE (RacE-V20) in the Y2H assay. Yeast was transformed with the indicated constructs and selected for the presence of prey and bait plasmids by growth on double-dropout (DD) media lacking leucine and tryptophan. Interactions were assayed by growth on stringent triple-dropout (TD) media additionally lacking histidine in the presence of 3 mM 3-amino-1,2,4-triazole (3-AT) and on quadruple-dropout (QD) media additionally lacking histidine and adenine. AD, Gal4-activation domain. (B) Genetic elimination of RacA and RacE was confirmed by immunoblotting. Porin was used as a loading control. (C) Constitutively active ForA fused to GFP requires RacE for targeting to the cell cortex but localizes appropriately in the absence of RacA. (Scale bars, 10 µm.) (D) ForA constructs used for biochemical analyses. Active RacE interacts directly with ForA-N and was able to partially release ForA-N from the autoinhibited ForA-N/ForA-C complex. GST-pulldown experiments with GMPPNP-loaded RacE are shown. The percentages below the lanes indicate the relative amounts of ForA-N in pellet (P) and supernatant (S) fractions. DD, dimerization domain; FH, formin homology domain. (E) Active RacE releases autoinhibition of the catalytically inactive ForA-N/ForA-C complex to promote actin assembly in pyrene assays in a concentration-dependent manner. (F) Active RacE N-terminally fused to YFP accumulates about twofold in the cell cortex of the cleavage furrow in 2D-confinement under agar. cf, cleavage furrow; p, pole. (Scale bar, 20 µm.) (G) Images from time-lapse movies correspond to Movie S1 and show that active RacE is enriched in the rear cell cortex of a polarized cell migrating under agar. (Scale bar, 20 µm.)